Knowles lab
The Knowles lab develops and applies machine learning methods for data analysis challenges in genomics. We're particularly interested in understanding the role of transcriptomic dysregulation, especially that involving RNA splicing, across the spectrum from rare to common genetic disease. This involves better characterization of the genetic and environmental factors contributing to mRNA expression and splicing variation. We collaborate with diverse research groups at NYGC and beyond collecting large-scale genomics datasets in the context of neurodegenerative and neuropsychiatric disease, and developing novel genomic technologies including single cell methods, forward genetic screens and long-read transcriptomics.
If you're interested in machine-learning-flavored genomics, statisical genetics, gene regulation and/or RNA splicing, please get in touch! We have experimental lab space so that includes wet lab people looking to do damp (mixed computational/experimental) projects. We're also interested in hearing from machine learning/statistics folks who are curious about applying their expertise in genetics/genomics. Postdocs and students are eligible for an appointment at Columbia University.
You can also browse old David's old personal website if you wish. David teaches COMS4762, a computer science class at Columbia on machine learning in genetics (Columbia LionMail account required for access), usually in the fall.
RNA splicing
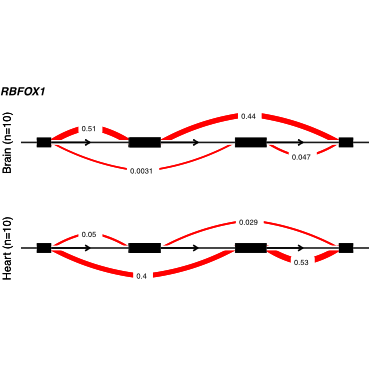
We developed LeafCutter to identify, quantify and test variable intron splicing events, obviating the need for accurate transcript annotations and circumventing the challenges in estimating relative isoform abundance (paper). An early version of LeafCutter was used in our study linking complex disease and splicing.
CRISPR/Cas13-based methods to study isoform function
A major challenge in characterizing the cellular function of different splice isoforms of a gene is the lack of methods to specifically and efficiently modulate their expression. We are currently developing Cas13-based strategies to 1) identify isoforms of the same gene with differential function in a high throughput manner and 2) to investigate the cellular functions of specific isoforms of interest.
Variant effects on RBP binding and splicing
Large sequencing consortia have identified thousands of genetic variants that are associated with splicing changes: splicing quantitative trait loci (sQTL). However, the causal variants and their intermediate molecular effects are largely unclear, although it is expected that many sQTLs function by altering binding of one or more RNA binding proteins (RBPs). We have several wet/dry lab projects trying to understand how non-coding variants affect RBP (especially splice factors) binding and, ultimately, splicing.
Causal inference in genetics
We're interested in designing statistical methods to infer causal relationships amongst traits and molecular phenotypes. For example, we developed Welch-weighted Egger regression to efficiently control for correlated pleiotropy (that is, a heritable confounding factor) in Mendelian randomization. We are working on approaches to infer direct causal effects from total causal effect estimates in high-dimensional settings.
Genetic differences in environmental response
Phenotypic variation results from the interplay of genotype and environment: genetic variation can modulate the transcriptomic response to perturbation. However, detecting gene by environment (GxE) effects on the transcriptome is challenging in observational data. We developed EAGLE to leverages allele-specific expression as a controlled, with-in individual test of the influence of environment factors on different genetic backgrounds. Another approach is direct in vitro perturbation of cells, which can greatly increase power to detect GxE due to increased effect sizes and controlled conditions. We used human iPSC-derived somatic cells to understand the genetic basis of anthracycline-cardiotoxicity (ACT), a common side-effect of chemotherapy. We measured transcriptomic and cellular damage response in a panel of iPSC-derived cardiomyocytes from 45 individuals. Using a novel, efficient linear mixed model, suez, we identified hundreds of loci that interact with ACT response and which are enriched in ACT GWAS (paper).
Variational methods
Variational methods offer a computationally efficient alternative to Markov chain Monte Carlo algorithms for inference in Bayesian probabilistic models. My work allows such methods to be more easily applied to a broader range of probabilistic models. I extended variational message passing to "non-conjugate" (intuitively, less tractable) models, and incorporated this method, Non-conjugate VMP (NCVMP), into the publicly available Infer.NET software package. Later, Tim Salimans and I did early work on using Monte Carlo estimation within variational learning, which is now an active subfield of research under the moniker "Stochastic Variational Inference".
Bayesian nonparametric models
Bayesian nonparametric (BNP) models are a category of statistical methods that automatically adapt their complexity to observed data. I have developed BNP methods for hierarchical clustering, heteroskedastic multivariate regression, network data, variable clustering and nonparametric sparse factor analysis (NSFA). In particular, NSFA is able to adaptively choose an appropriate number of factors from data. I used this method to delineate gene co-expression modules from microarray data, and other researchers have subsequently used it in diverse applications including image denoising and EEG analysis.
People
David A. Knowles, Principal Investigator
Staff
Chirag Lakhani - Staff Scientist

Chirag is broadly interested in utilizing methods from machine learning and statistical genetics to elucidate the biological function of genetic variants. Before joining NYGC he was a postdoctoral fellow in the Department of Biomedical Informatics at Harvard Medical School mentored by Chirag Patel. Before joining Harvard he worked in industry as a data scientist at Zaloni, Inc and HelloWallet. He received his PhD in mathematics at North Carolina State University. His undergraduate degree was also in mathematics from North Carolina State University. Outside of the lab he likes to spend time with his family either at home or exploring the city.
Saikat Banerjee - Staff Scientist

Saikat is passionate about understanding the regulatory mechanism of complex diseases. He specializes in developing statistical (Bayesian) methods and computational algorithms for analyzing large-scale multimodal NGS data. Currently, he focuses on studying the network effects of neuropsychiatric disorders. Before joining NYGC, he worked on developing fast Bayesian algorithms as a postdoctoral fellow in the group of Matthew Stephens at the University of Chicago. Previously, he worked at the Max Planck Institute for Multidisciplinary Sciences, Göttingen, with Johannes Söding, developing Bayesian methods for fine mapping and detecting trans-eQTLs. Saikat earned his Ph.D. in computational biophysics. Beyond his work, he enjoys history, philosophy, board games, fountain pen restoration, camping, hiking, and arthouse films. His personal website is here.
Jui-Shan (Teresa) Lin - Data Science Associate

Teresa joined the Knowles Lab in 2021. She is interested in statistical genetics and machine learning method to extract interpretable biological information from the human genome. Teresa completed her Master’s degree in the Bioinformatics and Genomics program at Penn State University. She worked on rare variant prioritization with Dr. Yifei Huang by integrating several supervised machine learning tools. Her undergraduate is in Biotechnology from National Cheng Kung University, Taiwan. Outside of work, Teresa is a foodie that likes to explore different cuisines. She also enjoys hiking or a nice jog along the Hudson River.
Claire Harbison - Associate Scientist

Claire joined the lab as a lab technician in 2022, after graduating with a BSc in chemistry and bioscience from the University of Pittsburgh. She has previously worked in neurobiological and clinical neurology labs, providing wet lab support to researchers. In the Knowles lab, she is assisting on multiple projects relating to alternative splicing and the impact of alternatively spliced isoforms on phenotype. She is also working to complete her Master's in Biomedical Laboratory Management at CUNY Hunter College.
Postdocs
Megan Schertzer

Megan joined the Knowles Lab as a postdoctoral research associate in 2019. She is interested in integrating human genomics and molecular biology to better understand the role of non-coding RNA in the brain. She originally completed a BS in biochemistry from Lee University in Tennessee, and obtained her PhD under the mentorship of Mauro Calabrese at UNC-Chapel Hill. There, she led a project to study how cis-acting long non-coding RNAs efficiently target and spread Polycomb Repressive Complexes to compact chromatin and repress nearby gene expression. Outside of the lab, she enjoys hiking, playing volleyball, and eating Mexican food.
Brielin C Brown

Brielin is a research fellow at the Data Science Institute, jointly affiliated with the New York Genome Center. Brielin's research lies at the intersection of machine learning and genomics. He is broadly interested in understanding how genetics and environment combine to lead to disease through changes in cellular function. To this end, he develops machine learning algorithms for modeling and inference in large-scale genomic studies. Before coming to Columbia, Brielin completed a PhD in Computer Science from the University of California, Berkeley and worked as a computational biologist at Verily Life Sciences. As a PhD student, Brielin was supported by an NSF fellowship and Chancellors fellowship for graduate study. He completed his undergraduate studies at the University of Virginia, earning a BS in Physics and a BA in Computer Science. In his spare time, Brielin enjoys techno music and is an avid surfer.
Scott Adamson

Scott joined the Knowles and Lappalainen labs in fall of 2021. He earned his BS in Environmental Science as well as his PhD in Biomedical Sciences from University of Connecticut. During his PhD, he worked under the supervision of Brenton Graveley to develop massively parallel reporter assays to identify the impact of genetic variance on pre-mRNA splicing, as well as understand splicing regulation more generally. In the Knowles lab, he is interested in developing new ways to predict and test the connection between genotype, splicing, and phenotype.
Aline Réal

Aline is interested in understanding the underlying mechanisms of gene expression regulation as a means to discovering and functionally interpreting genetic variants associated with splicing that contribute to human disease and traits. Aline was raised in the Aosta Valley, a small region of the Northern part of Italy. She completed her Master Degree in Molecular Biotechnology at the University of Turin and her PhD in genomic and digital health with Emmanouil Dermitzakis, Jörg Seebach and Ana Viñuela. There, she led two projects: the first using long-read direct-RNA-sequencing to identify specific variants associated with splicing and see their effect on transcript isoform expression and structure. With the second project she wanted to gain a deeper understanding of the regulatory germline contribution to cancer and to identify which target genes are involved using functional in-vitro screening assays. Outside of the lab, she enjoys cooking Italian dishes, skiing and playing violin.
Isabella Grabski

Isabella (Izzy) joined as a postdoctoral research associate in 2023, jointly with the Satija lab. She is interested in developing statistical and machine learning approaches to address challenges in single-cell genomics data analysis. She completed her PhD in Biostatistics at Harvard University, where she was advised by Professors Rafael Irizarry and Giovanni Parmigiani. Prior to her PhD, she completed her Bachelor's degree at Princeton University in Chemical and Biological Engineering. Outside of research, she enjoys hiking, reading, caring for her pet rats, and exploring the food scene of NYC.
Tatsuhiko Naito

Tatsuhiko joined the Knowles Lab and Raj Lab as a postdoctoral research fellow in 2023. His research interests lie in the application of machine learning to human genomes to gain a deeper understanding of the role of genetic variants in the pathogenesis of neurological diseases. He completed an MD and a PhD from the University of Tokyo in Japan and has experience working as a neurologist. During his PhD, he developed an HLA imputation method using deep learning and applied it to identify novel associations of HLA with diseases under the mentorship of Yukinori Okada at Osaka University. Additionally, he actively participated in human genetic research, including genome-wide association studies. Outside of the lab, he enjoys running in Central Park and working out at the gym.
Alex Tokolyi

Alex is interested in further understanding of the regulatory networks that connect common variation with transcriptional processes, and their downstream impacts on molecular phenotypes & disease. Alex grew up in Melbourne, Australia, completing undergraduate degrees at Monash University in Computer Science, Microbiology, and Molecular biology. During these studies he had the opportunity through a CSL and state-funded internship to pursue research at the Australian Regenerative Medicine Institute, creating systems to explore spatial transcriptomic data. Alex then performed research with Kathryn Holt in bacterial genomics, and Michael Inouye in asthma transcriptional networks, before moving to England to complete his PhD at the Sanger Institute in the University of Cambridge with Emma Davenport, assessing the genetic architecture of transcript splicing in healthy patients and those suffering sepsis.
PhD students
Andrew Stirn

Jiayu Su

Anjali Das

Karin Isaev

Sei Chang

Dan Meyer

Masters students
Xingpei Zhang

Ting Chen

Undergraduate students
Stella Park

Sophia Sowinski

Gill Bartels-Quansah

Alumni
Collin Wang

Siddhant (Sid) Sanghi

Kevin Wang

Peter Halmos - Data Science Associate

Peter recently completed his Bachelor’s degree at Columbia, majoring in Computer Science and concentrating in Chemistry. He is interested in means by which distinct forms of data can be used to inform priors and parameters which allow for the statistical quantification of allele specific binding of RNA binding proteins. He also has interests in Bayesian methods for fine-mapping and the learning of latent linear dynamical systems from discrete and sparse biological datasets. Previously, Peter was at Foundation Medicine’s Cancer Genomics Research division, and worked on both statistical methods and epidemiological projects for cancer genetics. In his free time, he enjoys hiking and nature photography.
Laura Pereira - Staff Scientist/Lab Manager

Chloé Terwagne

Stephen Malina

Nasrine Metic

Udai Nagpal

Join us!
We're always looking for great candidates to join the lab at all levels. If you're a student at Columbia University feel free to e-mail me about rotating in the lab. If you're looking at PhD programmes please bear in mind you will need to apply through Columbia University Computer Science or Systems Biology.
Publications
Working/under submission
- Stirn A and Knowles DA (2024), "The VampPrior Mixture Model"Abstract: Current clustering priors for deep latent variable models (DLVMs) require defining the number of clusters a-priori and are susceptible to poor initializations. Addressing these deficiencies could greatly benefit deep learning-based scRNA-seq analysis by performing integration and clustering simultaneously. We adapt the VampPrior (Tomczak & Welling, 2018) into a Dirichlet process Gaussian mixture model, resulting in the VampPrior Mixture Model (VMM), a novel prior for DLVMs. We propose an inference procedure that alternates between variational inference and Empirical Bayes to cleanly distinguish variational and prior parameters. Using the VMM in a Variational Autoencoder attains highly competitive clustering performance on benchmark datasets. Augmenting scVI (Lopez et al., 2018), a popular scRNA-seq integration method, with the VMM significantly improves its performance and automatically arranges cells into biologically meaningful clusters.BibTeX:
@article{Stirn2024-qw, author = {Stirn, Andrew and Knowles, David A}, title = {The VampPrior Mixture Model}, year = {2024}, url = {http://arxiv.org/abs/2402.04412} } - Park C, Mani S, Beltran-Velez N, Maurer K, Gohil S, Li S, Huang T, Knowles DA, Wu CJ and Azizi E (2023), "DIISCO: A Bayesian framework for inferring dynamic intercellular interactions from time-series single-cell data", RECOMB 2024 & under review at Genome Research.Abstract: Characterizing cell-cell communication and tracking its variability over time is essential for understanding the coordination of biological processes mediating normal development, progression of disease, or responses to perturbations such as therapies. Existing tools lack the ability to capture time-dependent intercellular interactions, such as those influenced by therapy, and primarily rely on existing databases compiled from limited contexts. We present DIISCO, a Bayesian framework for characterizing the temporal dynamics of cellular interactions using single-cell RNA-sequencing data from multiple time points. Our method uses structured Gaussian process regression to unveil time-resolved interactions among diverse cell types according to their co-evolution and incorporates prior knowledge of receptor-ligand complexes. We show the interpretability of DIISCO in simulated data and new data collected from CAR-T cells co-cultured with lymphoma cells, demonstrating its potential to uncover dynamic cell-cell crosstalk.BibTeX:
@article{Park2023-yh, author = {Park, Cameron and Mani, Shouvik and Beltran-Velez, Nicolas and Maurer, Katie and Gohil, Satyen and Li, Shuqiang and Huang, Teddy and Knowles, David A and Wu, Catherine J and Azizi, Elham}, title = {DIISCO: A Bayesian framework for inferring dynamic intercellular interactions from time-series single-cell data}, journal = {RECOMB 2024 & under review at Genome Research}, year = {2023}, url = {http://dx.doi.org/10.1101/2023.11.14.566956} } - Brown BC, Morris JA, Lappalainen T and Knowles DA (2023), "Large-scale causal discovery using interventional data sheds light on the regulatory network architecture of blood traits", bioRxiv. , pp. 2023.10.13.562293.Abstract: Inference of directed biological networks is an important but notoriously challenging problem. We introduce inverse sparse regression (inspre) , an approach to learning causal networks that leverages large-scale intervention-response data. Applied to 788 genes from the genome-wide perturb-seq dataset, inspre helps elucidate the network architecture of blood traits.BibTeX:
@article{Brown2023-uc, author = {Brown, Brielin C and Morris, John A and Lappalainen, Tuuli and Knowles, David A}, title = {Large-scale causal discovery using interventional data sheds light on the regulatory network architecture of blood traits}, journal = {bioRxiv}, year = {2023}, pages = {2023.10.13.562293}, url = {https://www.biorxiv.org/content/10.1101/2023.10.13.562293v1} } - Schertzer MD, Stirn A, Isaev K, Pereira L, Das A, Harbison C, Park SH, Wessels H-H, Sanjana NE and Knowles DA (2023), "Cas13d-mediated isoform-specific RNA knockdown with a unified computational and experimental toolbox", bioRxiv. , pp. 2023.09.12.557474.Abstract: Alternative splicing is an essential mechanism for diversifying proteins, in which mature RNA isoforms produce proteins with potentially distinct functions. Two major challenges in characterizing the cellular function of isoforms are the lack of experimental methods to specifically and efficiently modulate isoform expression and computational tools for complex experimental design. To address these gaps, we developed and methodically tested a strategy which pairs the RNA-targeting CRISPR/Cas13d system with guide RNAs that span exon-exon junctions in the mature RNA. We performed a high-throughput essentiality screen, quantitative RT-PCR assays, and PacBio long read sequencing to affirm our ability to specifically target and robustly knockdown individual RNA isoforms. In parallel, we provide computational tools for experimental design and screen analysis. Considering all possible splice junctions annotated in GENCODE for multi-isoform genes and our gRNA efficacy predictions, we estimate that our junction-centric strategy can uniquely target up to 89% of human RNA isoforms, including 50,066 protein-coding and 11,415 lncRNA isoforms. Importantly, this specificity spans all splicing and transcriptional events, including exon skipping and inclusion, alternative 5' and 3' splice sites, and alternative starts and ends.BibTeX:
@article{Schertzer2023-lb, author = {Schertzer, Megan D and Stirn, Andrew and Isaev, Keren and Pereira, Laura and Das, Anjali and Harbison, Claire and Park, Stella H and Wessels, Hans-Hermann and Sanjana, Neville E and Knowles, David A}, title = {Cas13d-mediated isoform-specific RNA knockdown with a unified computational and experimental toolbox}, journal = {bioRxiv}, year = {2023}, pages = {2023.09.12.557474}, url = {https://www.biorxiv.org/content/10.1101/2023.09.12.557474v1} } - Su J, Reynier J-B, Fu X, Zhong G, Jiang J, Escalante RS, Wang Y, Izar B, Knowles DA and Rabadan R (2022), "A Unified Modular Framework to Incorporate Structural Dependency in Spatial Omics Data", bioRxiv. , pp. 2022.10.25.513785.Abstract: Spatial omics technologies, such as spatial transcriptomics, allow the identification of spatially organized biological processes, while presenting computational challenges for existing analysis approaches that ignore spatial dependencies. Here we introduce Smoother, a unified and modular framework that integrates positional information into non-spatial models via spatial priors and losses. In simulated and real datasets, we show that Smoother enables spatially aware data imputation, cell-type deconvolution, and dimensionality reduction with high accuracy.BibTeX:
@article{Su2022, author = {Su, Jiayu and Reynier, Jean-Baptiste and Fu, Xi and Zhong, Guojie and Jiang, Jiahao and Escalante, Rydberg Supo and Wang, Yiping and Izar, Benjamin and Knowles, David A and Rabadan, Raul}, title = {A Unified Modular Framework to Incorporate Structural Dependency in Spatial Omics Data}, journal = {bioRxiv}, year = {2022}, pages = {2022.10.25.513785}, url = {https://www.biorxiv.org/content/10.1101/2022.10.25.513785v1.abstract} } - Brown BC and Knowles DA (2020), "Phenome-scale causal network discovery with bidirectional mediated Mendelian randomization", bioRxiv.Abstract: Inference of directed biological networks from observational genomics datasets is a crucial but notoriously difficult challenge. Modern population-scale biobanks, containing simultaneous measurements of traits, biomarkers, and genetic variation, offer an unprecedented opportunity to study biological networks. Mendelian randomization (MR) has received attention as a class of methods for inferring causal effects in observational data that uses genetic variants as instrumental variables, but MR methods rely on assumptions that limit their application to complex traits at the biobank-scale. Moreover, MR estimates the total effect of one trait on another, which may be mediated by other factors. Biobanks include measurements of many potential mediators, in principle enabling the conversion of MR estimates into direct effects representing a causal network. Here, we show that this can be accomplished by a flexible two stage procedure we call bidirectional mediated Mendelian randomization (bimmer). First, bimmer estimates the effect of every trait on every other. Next, bimmer finds a parsimonious network that explains these effects using direct and mediated causal paths. We introduce novel methods for both steps and show via extensive simulations that bimmer is able to learn causal network structures even in the presence of non-causal genetic correlation. We apply bimmer to 405 phenotypes from the UK biobank and demonstrate that learning the network structure is invaluable for interpreting the results of phenome-wide MR, while lending causal support to several recent observational studies.BibTeX:
@article{Brown2020, author = {Brown, Brielin C. and Knowles, David A.}, title = {Phenome-scale causal network discovery with bidirectional mediated Mendelian randomization}, journal = {bioRxiv}, publisher = {Cold Spring Harbor Laboratory}, year = {2020}, url = {https://www.biorxiv.org/content/early/2020/06/22/2020.06.18.160176}, doi = {10.1101/2020.06.18.160176} } - Stirn A and Knowles DA (2020), "Variational Variance: Simple and Reliable Predictive Variance Parameterization", arXiv.Abstract: Brittle optimization has been observed to adversely impact model likelihoods for regression and VAEs when simultaneously fitting neural network mappings from a (random) variable onto the mean and variance of a dependent Gaussian variable. Previous works have bolstered optimization and improved likelihoods, but fail other basic posterior predictive checks (PPCs). Under the PPC framework, we propose critiques to test predictive mean and variance calibration and the predictive distribution's ability to generate sensible data. We find that our attractively simple solution, to treat heteroscedastic variance variationally, sufficiently regularizes variance to pass these PPCs. We consider a diverse gamut of existing and novel priors and find our methods preserve or outperform existing model likelihoods while significantly improving parameter calibration and sample quality for regression and VAEs.BibTeX:
@article{stirn2020variational, author = {Stirn, Andrew and Knowles, David A}, title = {Variational Variance: Simple and Reliable Predictive Variance Parameterization}, journal = {arXiv}, year = {2020}, url = {https://arxiv.org/abs/2006.04910}, doi = {arXiv:2006.04910v2} } - Knowles DA (2015), "Stochastic gradient variational Bayes for gamma approximating distributions", arXiv. , pp. 1509.01631.Abstract: While stochastic variational inference is relatively well known for scaling inference in Bayesian probabilistic models, related methods also offer ways to circumnavigate the approximation of analytically intractable expectations. The key challenge in either setting is controlling the variance of gradient estimates: recent work has shown that for continuous latent variables, particularly multivariate Gaussians, this can be achieved by using the gradient of the log posterior. In this paper we apply the same idea to gamma distributed latent variables given gamma variational distributions, enabling straightforward "black box" variational inference in models where sparsity and non-negativity are appropriate. We demonstrate the method on a recently proposed gamma process model for network data, as well as a novel sparse factor analysis. We outperform generic sampling algorithms and the approach of using Gaussian variational distributions on transformed variables.BibTeX:
@article{Knowles2015stochastic, author = {Knowles, David A}, title = {Stochastic gradient variational Bayes for gamma approximating distributions}, journal = {arXiv}, year = {2015}, pages = {1509.01631}, url = {https://arxiv.org/abs/1509.01631} } - Salimans T and Knowles DA (2014), "On using control variates with stochastic approximation for variational Bayes and its connection to stochastic linear regression", arXiv. , pp. 1401.1022.Abstract: Recently, we and several other authors have written about the possibilities of using stochastic approximation techniques for fitting variational approximations to intractable Bayesian posterior distributions. Naive implementations of stochastic approximation suffer from high variance in this setting. Several authors have therefore suggested using control variates to reduce this variance, while we have taken a different but analogous approach to reducing the variance which we call stochastic linear regression. In this note we take the former perspective and derive the ideal set of control variates for stochastic approximation variational Bayes under a certain set of assumptions. We then show that using these control variates is closely related to using the stochastic linear regression approximation technique we proposed earlier. A simple example shows that our method for constructing control variates leads to stochastic estimators with much lower variance compared to other approaches.BibTeX:
@article{salimans2014using, author = {Salimans, Tim and Knowles, David A}, title = {On using control variates with stochastic approximation for variational Bayes and its connection to stochastic linear regression}, journal = {arXiv}, year = {2014}, pages = {1401.1022}, url = {https://arxiv.org/abs/1401.1022} } - Palla* K, Knowles* DA and Ghahramani Z (2013), "A dependent partition-valued process for multitask clustering and time evolving network modeling", arXiv. , pp. 1303.3265. *These authors contributed equally to this work.Abstract: The fundamental aim of clustering algorithms is to partition data points. We consider tasks where the discovered partition is allowed to vary with some covariate such as space or time. One approach would be to use fragmentation-coagulation processes, but these, being Markov processes, are restricted to linear or tree structured covariate spaces. We define a partition-valued process on an arbitrary covariate space using Gaussian processes. We use the process to construct a multitask clustering model which partitions datapoints in a similar way across multiple data sources, and a time series model of network data which allows cluster assignments to vary over time. We describe sampling algorithms for inference and apply our method to defining cancer subtypes based on different types of cellular characteristics, finding regulatory modules from gene expression data from multiple human populations, and discovering time varying community structure in a social network.BibTeX:
@article{palla2013dependent, author = {Palla*, Konstantina and Knowles*, David A and Ghahramani, Zoubin}, title = {A dependent partition-valued process for multitask clustering and time evolving network modeling}, journal = {arXiv}, year = {2013}, pages = {1303.3265}, url = {https://arxiv.org/abs/1303.3265} }
Genetics
- Su J, Reynier J-B, Fu X, Zhong G, Jiang J, Escalante RS, Wang Y, Aparicio L, Izar B, Knowles DA and Rabadan R (2023), "Smoother: a unified and modular framework for incorporating structural dependency in spatial omics data", Genome biology. Vol. 24(1), pp. 291.BibTeX:
@article{Su2023, author = {Su, Jiayu and Reynier, Jean-Baptiste and Fu, Xi and Zhong, Guojie and Jiang, Jiahao and Escalante, Rydberg Supo and Wang, Yiping and Aparicio, Luis and Izar, Benjamin and Knowles, David A and Rabadan, Raul}, title = {Smoother: a unified and modular framework for incorporating structural dependency in spatial omics data}, journal = {Genome biology}, year = {2023}, volume = {24}, number = {1}, pages = {291}, url = {http://dx.doi.org/10.1186/s13059-023-03138-x} } - Isaev K and Knowles DA (2023), "Investigating RNA splicing as a source of cellular diversity using a binomial mixture model", In Proceedings of Machine Learning Research: MLCB 2023.Abstract: Alternative splicing (AS) contributes significantly to RNA and protein variability yet its role in defining cellular diversity is not fully understood. While Smart-seq2 offers enhanced coverage across transcripts compared to 10X single cell RNA-sequencing (scRNA-seq), current computational methods often miss the full complexity of AS. Most approaches for single cell based differential splicing analysis focus on simple AS events such as exon skipping, and rely on predefined cell type labels or low-dimensional gene expression representations. This limits their ability to detect more complex AS events and makes them dependent on prior knowledge of cell classifications. Here, we present Leaflet, a splice junction centric approach inspired by Leafcutter, our tool for quantifying RNA splicing variation with bulk RNA-seq. Leaflet is a probabilistic mixture model designed to infer AS-driven cell states without the need for cell type labels. We detail Leaflet’s generative model, inference methodology, and its efficiency in detecting differentially spliced junctions. By applying Leaflet to the Tabula Muris brain cell dataset, we highlight cell-state specific splicing patterns, offering a deeper insight into cellular diversity beyond that captured by gene expression alone.BibTeX:
@inproceedings{Isaev2023-ax, author = {Isaev, Keren and Knowles, David A}, title = {Investigating RNA splicing as a source of cellular diversity using a binomial mixture model}, booktitle = {Proceedings of Machine Learning Research: MLCB 2023}, year = {2023}, url = {https://proceedings.mlr.press/v240/isaev24a.html} } - Wessels* H-H, Stirn* A, Méndez-Mancilla A, Kim EJ, Hart SK, Knowles† DA and Sanjana† NE (2023), "Prediction of on-target and off-target activity of CRISPR-Cas13d guide RNAs using deep learning", Nature biotechnology. *Equal contribution. †Co-corresponding.Abstract: Transcriptome engineering applications in living cells with RNA-targeting CRISPR effectors depend on accurate prediction of on-target activity and off-target avoidance. Here we design and test 200,000 RfxCas13d guide RNAs targeting essential genes in human cells with systematically designed mismatches and insertions and deletions (indels). We find that mismatches and indels have a position- and context-dependent impact on Cas13d activity, and mismatches that result in G--U wobble pairings are better tolerated than other single-base mismatches. Using this large-scale dataset, we train a convolutional neural network that we term targeted inhibition of gene expression via gRNA design (TIGER) to predict efficacy from guide sequence and context. TIGER outperforms the existing models at predicting on-target and off-target activity on our dataset and published datasets. We show that TIGER scoring combined with specific mismatches yields the first general framework to modulate transcript expression, enabling the use of RNA-targeting CRISPRs to precisely control gene dosage. A machine learning model predicts on-target and off-target activity of Cas13d in human cells.BibTeX:
@article{Wessels2023-ko, author = {Wessels*, Hans-Hermann and Stirn*, Andrew and Méndez-Mancilla, Alejandro and Kim, Eric J and Hart, Sydney K and Knowles†, David A and Sanjana†, Neville E}, title = {Prediction of on-target and off-target activity of CRISPR-Cas13d guide RNAs using deep learning}, journal = {Nature biotechnology}, year = {2023}, url = {https://www.nature.com/articles/s41587-023-01830-8} } - Cortés-López M, Chamely P, Hawkins AG, Stanley RF, Swett AD, Ganesan S, Mouhieddine TH, Dai X, Kluegel L, Chen C, Batta K, Furer N, Vedula RS, Beaulaurier J, Drong AW, Hickey S, Dusaj N, Mullokandov G, Stasiw AM, Su J, Chaligné R, Juul S, Harrington E, Knowles DA, Potenski CJ, Wiseman DH, Tanay A, Shlush L, Lindsley RC, Ghobrial IM, Taylor J, Abdel-Wahab O, Gaiti F and Landau DA (2023), "Single-cell multi-omics defines the cell-type-specific impact of splicing aberrations in human hematopoietic clonal outgrowths", Cell Stem Cell.Abstract: Summary RNA splicing factors are recurrently mutated in clonal blood disorders, but the impact of dysregulated splicing in hematopoiesis remains unclear. To overcome technical limitations, we integrated genotyping of transcriptomes (GoT) with long-read single-cell transcriptomics and proteogenomics for single-cell profiling of transcriptomes, surface proteins, somatic mutations, and RNA splicing (GoT-Splice). We applied GoT-Splice to hematopoietic progenitors from myelodysplastic syndrome (MDS) patients with mutations in the core splicing factor SF3B1. SF3B1mut cells were enriched in the megakaryocytic-erythroid lineage, with expansion of SF3B1mut erythroid progenitor cells. We uncovered distinct cryptic 3′ splice site usage in different progenitor populations and stage-specific aberrant splicing during erythroid differentiation. Profiling SF3B1-mutated clonal hematopoiesis samples revealed that erythroid bias and cell-type-specific cryptic 3′ splice site usage in SF3B1mut cells precede overt MDS. Collectively, GoT-Splice defines the cell-type-specific impact of somatic mutations on RNA splicing, from early clonal outgrowths to overt neoplasia, directly in human samples.BibTeX:
@article{Cortes-Lopez2023-sk, author = {Cortés-López, Mariela and Chamely, Paulina and Hawkins, Allegra G and Stanley, Robert F and Swett, Ariel D and Ganesan, Saravanan and Mouhieddine, Tarek H and Dai, Xiaoguang and Kluegel, Lloyd and Chen, Celine and Batta, Kiran and Furer, Nili and Vedula, Rahul S and Beaulaurier, John and Drong, Alexander W and Hickey, Scott and Dusaj, Neville and Mullokandov, Gavriel and Stasiw, Adam M and Su, Jiayu and Chaligné, Ronan and Juul, Sissel and Harrington, Eoghan and Knowles, David A and Potenski, Catherine J and Wiseman, Daniel H and Tanay, Amos and Shlush, Liran and Lindsley, Robert C and Ghobrial, Irene M and Taylor, Justin and Abdel-Wahab, Omar and Gaiti, Federico and Landau, Dan A}, title = {Single-cell multi-omics defines the cell-type-specific impact of splicing aberrations in human hematopoietic clonal outgrowths}, journal = {Cell Stem Cell}, year = {2023}, url = {https://www.sciencedirect.com/science/article/pii/S1934590923002576} } - Brown* BC, Wang C, Kasela S, Aguet F, Nachun DC, Taylor KD, Tracy RP, Durda P, Liu Y, Craig Johnson W, Van Den Berg D, Gupta N, Gabriel S, Smith JD, Gerzsten R, Clish C, Wong Q, Papanicolau G, Blackwell TW, Rotter JI, Rich SS, Graham Barr R, Ardlie KG, Knowles* DA and Lappalainen* T (2023), "Multiset correlation and factor analysis enables exploration of multi-omics data", Cell Genomics. *Co-corresponding.Abstract: Multi-omics datasets are becoming more common, necessitating better integration methods to realize their revolutionary potential. Here, we introduce multi-set correlation and factor analysis (MCFA), an unsupervised integration method tailored to the unique challenges of high-dimensional genomics data that enables fast inference of shared and private factors. We used MCFA to integrate methylation markers, protein expression, RNA expression, and metabolite levels in 614 diverse samples from the Trans-Omics for Precision Medicine/Multi-Ethnic Study of Atherosclerosis multi-omics pilot. Samples cluster strongly by ancestry in the shared space, even in the absence of genetic information, while private spaces frequently capture dataset-specific technical variation. Finally, we integrated genetic data by conducting a genome-wide association study (GWAS) of our inferred factors, observing that several factors are enriched for GWAS hits and trans-expression quantitative trait loci. Two of these factors appear to be related to metabolic disease. Our study provides a foundation and framework for further integrative analysis of ever larger multi-modal genomic datasets.BibTeX:
@article{Brown2023-fm, author = {Brown*, Brielin C and Wang, Collin and Kasela, Silva and Aguet, Fran¸ cois and Nachun, Daniel C and Taylor, Kent D and Tracy, Russell P and Durda, Peter and Liu, Yongmei and Craig Johnson, W and Van Den Berg, David and Gupta, Namrata and Gabriel, Stacy and Smith, Joshua D and Gerzsten, Robert and Clish, Clary and Wong, Quenna and Papanicolau, George and Blackwell, Thomas W and Rotter, Jerome I and Rich, Stephen S and Graham Barr, R and Ardlie, Kristin G and Knowles*, David A and Lappalainen*, Tuuli}, title = {Multiset correlation and factor analysis enables exploration of multi-omics data}, journal = {Cell Genomics}, year = {2023}, url = {http://www.cell.com/article/S2666979X23001428/abstract} } - Malina S, Cizin D and Knowles DA (2022), "Deep Mendelian randomization: Investigating the causal knowledge of genomic deep learning models", PLOS Computational Biology. Vol. 18(10), pp. 1-14.Abstract: Multi-task deep learning (DL) models can accurately predict diverse genomic marks from sequence, but whether these models learn the causal relationships between genomic marks is unknown. Here, we describe Deep Mendelian Randomization (DeepMR), a method for estimating causal relationships between genomic marks learned by genomic DL models. By combining Mendelian randomization with in silico mutagenesis, DeepMR obtains local (locus specific) and global estimates of (an assumed) linear causal relationship between marks. In a simulation designed to test recovery of pairwise causal relations between transcription factors (TFs), DeepMR gives accurate and unbiased estimates of the true global causal effect, but its coverage decays in the presence of sequence-dependent confounding. We then apply DeepMR to examine the global relationships learned by a state-of-the-art DL model, BPNet, between TFs involved in reprogramming. DeepMR's causal effect estimates validate previously hypothesized relationships between TFs and suggest new relationships for future investigation.BibTeX:
@article{Malina2022, author = {Malina, Stephen AND Cizin, Daniel AND Knowles, David A.}, title = {Deep Mendelian randomization: Investigating the causal knowledge of genomic deep learning models}, journal = {PLOS Computational Biology}, publisher = {Public Library of Science}, year = {2022}, volume = {18}, number = {10}, pages = {1-14}, url = {https://doi.org/10.1371/journal.pcbi.1009880}, doi = {10.1371/journal.pcbi.1009880} } - Humphrey J, Venkatesh S, Hasan R, Herb JT, de Paiva Lopes K, Kü¸ cükali F, Byrska-Bishop M, Evani US, Narzisi G, Fagegaltier D, NYGC ALS Consortium, Sleegers K, Phatnani H, Knowles DA, Fratta P and Raj T (2022), "Integrative transcriptomic analysis of the amyotrophic lateral sclerosis spinal cord implicates glial activation and suggests new risk genes", Nature Neuroscience.Abstract: Amyotrophic lateral sclerosis (ALS) is a progressively fatal neurodegenerative disease affecting motor neurons in the brain and spinal cord. In this study, we investigated gene expression changes in ALS via RNA sequencing in 380 postmortem samples from cervical, thoracic and lumbar spinal cord segments from 154 individuals with ALS and 49 control individuals. We observed an increase in microglia and astrocyte gene expression, accompanied by a decrease in oligodendrocyte gene expression. By creating a gene co-expression network in the ALS samples, we identified several activated microglia modules that negatively correlate with retrospective disease duration. We mapped molecular quantitative trait loci and found several potential ALS risk loci that may act through gene expression or splicing in the spinal cord and assign putative cell types for FNBP1, ACSL5, SH3RF1 and NFASC. Finally, we outline how common genetic variants associated with splicing of C9orf72 act as proxies for the well-known repeat expansion, and we use the same mechanism to suggest ATXN3 as a putative risk gene.BibTeX:
@article{Humphrey2022, author = {Humphrey, Jack and Venkatesh, Sanan and Hasan, Rahat and Herb, Jake T and de Paiva Lopes, Katia and Kü¸ cükali, Fahri and Byrska-Bishop, Marta and Evani, Uday S and Narzisi, Giuseppe and Fagegaltier, Delphine and NYGC ALS Consortium and Sleegers, Kristel and Phatnani, Hemali and Knowles, David A and Fratta, Pietro and Raj, Towfique}, title = {Integrative transcriptomic analysis of the amyotrophic lateral sclerosis spinal cord implicates glial activation and suggests new risk genes}, journal = {Nature Neuroscience}, year = {2022}, url = {https://www.medrxiv.org/content/10.1101/2021.08.31.21262682v1} } - Brown BC and Knowles DA (2021), "Welch-weighted Egger regression reduces false positives due to correlated pleiotropy in Mendelian randomization", American Journal of Human Genetics.Abstract: Modern population-scale biobanks contain simultaneous measurements of many phenotypes, providing unprecedented opportunity to study the relationship between biomarkers and disease. However, inferring causal effects from observational data is notoriously challenging. Mendelian randomization (MR) has recently received increased attention as a class of methods for estimating causal effects using genetic associations. However, standard methods result in pervasive false positives when two traits share a heritable, unobserved common cause. This is the problem of correlated pleiotropy. Here, we introduce a flexible framework for simulating traits with a common genetic confounder that generalizes recently proposed models, as well as a simple approach we call Welch-weighted Egger regression (WWER) for estimating causal effects. We show in comprehensive simulations that our method substantially reduces false positives due to correlated pleiotropy while being fast enough to apply to hundreds of phenotypes. We apply our method first to a subset of the UK Biobank consisting of blood traits and inflammatory disease, and then to a broader set of 411 heritable phenotypes. We detect many effects with strong literature support, as well as numerous behavioral effects that appear to stem from physician advice given to people at high risk for disease. We conclude that WWER is a powerful tool for exploratory data analysis in ever-growing databases of genotypes and phenotypes.BibTeX:
@article{Brown2021, author = {Brown, Brielin C and Knowles, David A}, title = {Welch-weighted Egger regression reduces false positives due to correlated pleiotropy in Mendelian randomization}, journal = {American Journal of Human Genetics}, year = {2021}, url = {https://www.cell.com/ajhg/fulltext/S0002-9297(21)00383-9} } - Hadi K, Yao X, Behr JM, Deshpande A, Xanthopoulakis C, Rosiene J, Darmofal M, Tian H, DeRose J, Mortensen R, Adney EM, Gajic Z, Eng K, Wala JA, Wrzeszczyʼnski KO, Arora K, Shah M, Emde A-K, Felice V, Frank MO, Darnell RB, Ghandi M, Huang F, Maciejowski J, De Lange T, Setton J, Riaz N, Reis-Filho JS, Powell S, Knowles D, Reznik E, Mishra B, Beroukhim R, Zody M, Robine N, Oman KM, Sanchez CA, Kuhner MK, Smith LP, Galipeau PC, Paulson TG, Reid BJ, Li X, Wilkes D, Sboner A, Mosquera JM, Elemento O and Imielinski M (2020), "Novel patterns of complex structural variation revealed across thousands of cancer genome graphs", Cell.Abstract: Cancer genomes often harbor hundreds of somatic DNA rearrangement junctions, many of which cannot be easily classified into simple (e.g. deletion, translocation) or complex (e.g. chromothripsis, chromoplexy) structural variant classes. Applying a novel genome graph computational paradigm to analyze the topology of junction copy number (JCN) across 2,833 tumor whole genome sequences (WGS), we introduce three complex rearrangement phenomena: pyrgo, rigma, and tyfonas. Pyrgo are “towers” of low-JCN duplications associated with early replicating regions and superenhancers, and are enriched in breast and ovarian cancers. Rigma comprise “chasms” of low-JCN deletions at late-replicating fragile sites in esophageal and other gastrointestinal (GI) adenocarcinomas. Tyfonas are “typhoons” of high-JCN junctions and fold back inversions that are enriched in acral but not cutaneous melanoma and associated with a previously uncharacterized mutational process of non-APOBEC kataegis. Clustering of tumors according to genome graph-derived features identifies subgroups associated with DNA repair defects and poor prognosis.BibTeX:
@article{Hadi2020, author = {Hadi, Kevin and Yao, Xiaotong and Behr, Julie M. and Deshpande, Aditya and Xanthopoulakis, Charalampos and Rosiene, Joel and Darmofal, Madison and Tian, Huasong and DeRose, Joseph and Mortensen, Rick and Adney, Emily M. and Gajic, Zoran and Eng, Kenneth and Wala, Jeremiah A. and Wrzeszczyʼnski, Kazimierz O. and Arora, Kanika and Shah, Minita and Emde, Anne-Katrin and Felice, Vanessa and Frank, Mayu O. and Darnell, Robert B. and Ghandi, Mahmoud and Huang, Franklin and Maciejowski, John and De Lange, Titia and Setton, Jeremy and Riaz, Nadeem and Reis-Filho, Jorge S. and Powell, Simon and Knowles, David and Reznik, Ed and Mishra, Bud and Beroukhim, Rameen and Zody, Michael and Robine, Nicolas and Oman, Kenji M. and Sanchez, Carissa A. and Kuhner, Mary K. and Smith, Lucian P. and Galipeau, Patricia C. and Paulson, Thomas G. and Reid, Brian J. and Li, Xiaohong and Wilkes, David and Sboner, Andrea and Mosquera, Juan Miguel and Elemento, Olivier and Imielinski, Marcin}, title = {Novel patterns of complex structural variation revealed across thousands of cancer genome graphs}, journal = {Cell}, year = {2020}, url = {https://www.cell.com/cell/pdf/S0092-8674(20)30997-1.pdf}, doi = {10.1016/j.cell.2020.08.006} } - Gentles AJ, Hui AB-Y, Feng W, Azizi A, Nair RV, Knowles DA, Yu A, Jeong Y, Bejnood A, Forgó E, Varma S, Xu Y, Kuong A, Nair VS, West R, van de Rijn M, Hoang CD, Diehn M and Plevritis SK (2020), "Clinically-relevant cell type cross-talk identified from a human lung tumor microenvironment interactome", Genome Biology.Abstract: Tumors comprise a complex microenvironment of interacting malignant and stromal cell types. Much of our understanding of the tumor microenvironment comes from in vitro studies isolating the interactions between malignant cells and a single stromal cell type, often along a single pathway. To develop a deeper understanding of the interactions between cells within human lung tumors we performed RNA-seq profiling of flow-sorted malignant cells, endothelial cells, immune cells, fibroblasts, and bulk cells from freshly resected human primary non-small-cell lung tumors. We mapped the cell-specific differential expression of prognostically-associated secreted factors and cell surface genes, and computationally reconstructed cross-talk between these cell types to generate a novel resource we call the Lung Tumor Microenvironment Interactome (LTMI). Using this resource, we identified and validated a prognostically unfavorable influence of Gremlin-1 production by fibroblasts on proliferation of malignant lung adenocarcinoma cells. We also found a prognostically favorable association between infiltration of mast cells and less aggressive tumor cell behavior. These results illustrate the utility of the LTMI as a resource for generating hypotheses concerning tumor-microenvironment interactions that may have prognostic and therapeutic relevance. Summary RNA-seq profiling of sorted populations from primary lung cancer samples identifies prognostically relevant cross-talk between cell types in the tumor microenvironment.BibTeX:
@article{Gentles2019, author = {Gentles, Andrew J and Hui, Angela Bik-Yu and Feng, Weiguo and Azizi, Armon and Nair, Ramesh V. and Knowles, David A. and Yu, Alice and Jeong, Youngtae and Bejnood, Alborz and Forgó, Erna and Varma, Sushama and Xu, Yue and Kuong, Amanda and Nair, Viswam S. and West, Rob and van de Rijn, Matt and Hoang, Chuong D. and Diehn, Maximilian and Plevritis, Sylvia K.}, title = {Clinically-relevant cell type cross-talk identified from a human lung tumor microenvironment interactome}, journal = {Genome Biology}, year = {2020}, url = {https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-02019-x}, doi = {10.1101/637306} } - Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, Kosmicki JA, Arbelaez J, Cui W, Schwartz GB, Chow ED, Kanterakis E, Gao H, Kia A, Batzoglou S, Sanders SJ and Farh KK-H (2019), "Predicting Splicing from Primary Sequence with Deep Learning", Cell. Vol. 176(3), pp. 535-548.e24.Abstract: The splicing of pre-mRNAs into mature transcripts is remarkable for its precision, but the mechanisms by which the cellular machinery achieves such specificity are incompletely understood. Here, we describe a deep neural network that accurately predicts splice junctions from an arbitrary pre-mRNA transcript sequence, enabling precise prediction of noncoding genetic variants that cause cryptic splicing. Synonymous and intronic mutations with predicted splice-altering consequence validate at a high rate on RNA-seq and are strongly deleterious in the human population. De novo mutations with predicted splice-altering consequence are significantly enriched in patients with autism and intellectual disability compared to healthy controls and validate against RNA-seq in 21 out of 28 of these patients. We estimate that 9%-11% of pathogenic mutations in patients with rare genetic disorders are caused by this previously underappreciated class of disease variation.BibTeX:
@article{Jaganathan2019-kb, author = {Jaganathan, Kishore and Kyriazopoulou Panagiotopoulou, Sofia and McRae, Jeremy F and Darbandi, Siavash Fazel and Knowles, David and Li, Yang I and Kosmicki, Jack A and Arbelaez, Juan and Cui, Wenwu and Schwartz, Grace B and Chow, Eric D and Kanterakis, Efstathios and Gao, Hong and Kia, Amirali and Batzoglou, Serafim and Sanders, Stephan J and Farh, Kyle Kai-How}, title = {Predicting Splicing from Primary Sequence with Deep Learning}, journal = {Cell}, year = {2019}, volume = {176}, number = {3}, pages = {535--548.e24} } - Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, Ermel R, Ruusalepp A, Quertermous T, Hao K, Björkegren JLM, Im HK, Pasaniuc B, Rivas MA and Kundaje A (2019), "Opportunities and challenges for transcriptome-wide association studies", Nature Genetics. Vol. 51(4), pp. 592-599.Abstract: Transcriptome-wide association studies (TWAS) integrate genome-wide association studies (GWAS) and gene expression datasets to identify gene-trait associations. In this Perspective, we explore properties of TWAS as a potential approach to prioritize causal genes at GWAS loci, by using simulations and case studies of literature-curated candidate causal genes for schizophrenia, low-density-lipoprotein cholesterol and Crohn's disease. We explore risk loci where TWAS accurately prioritizes the likely causal gene as well as loci where TWAS prioritizes multiple genes, some likely to be non-causal, owing to sharing of expression quantitative trait loci (eQTL). TWAS is especially prone to spurious prioritization with expression data from non-trait-related tissues or cell types, owing to substantial cross-cell-type variation in expression levels and eQTL strengths. Nonetheless, TWAS prioritizes candidate causal genes more accurately than simple baselines. We suggest best practices for causal-gene prioritization with TWAS and discuss future opportunities for improvement. Our results showcase the strengths and limitations of using eQTL datasets to determine causal genes at GWAS loci.BibTeX:
@article{Wainberg2019-lb, author = {Wainberg, Michael and Sinnott-Armstrong, Nasa and Mancuso, Nicholas and Barbeira, Alvaro N and Knowles, David A and Golan, David and Ermel, Raili and Ruusalepp, Arno and Quertermous, Thomas and Hao, Ke and Björkegren, Johan L M and Im, Hae Kyung and Pasaniuc, Bogdan and Rivas, Manuel A and Kundaje, Anshul}, title = {Opportunities and challenges for transcriptome-wide association studies}, journal = {Nature Genetics}, year = {2019}, volume = {51}, number = {4}, pages = {592--599} } - Calderon D, Nguyen ML, Mezger A, Kathiria A, Nguyen V, Lescano N, Wu B, Trombetta J, Ribado JV, Knowles DA, Gao Z, Parent AV, Burt TD, Anderson MS, Criswell LA, Greenleaf WJ, Marson A and Pritchard JK (2019), "Landscape of stimulation-responsive chromatin across diverse human immune cells", Nature Genetics.Abstract: The immune system is controlled by a balanced interplay among specialized cell types transitioning between resting and stimulated states. Despite its importance, the regulatory landscape of this system has not yet been fully characterized. To address this gap, we collected ATAC-seq and RNA-seq data under resting and stimulated conditions for 25 immune cell types from peripheral blood of four healthy individuals, and seven cell types from three fetal thymus samples. We found that stimulation caused widespread chromatin remodeling, including a large class of response elements shared between stimulated B and T cells. Furthermore, several autoimmune traits showed significant heritability in stimulation-responsive elements from distinct cell types, highlighting the critical importance of these cell states in autoimmunity. Use of allele-specific read-mapping identified thousands of variants that alter chromatin accessibility in particular conditions. Notably, variants associated with changes in stimulation-specific chromatin accessibility were not enriched for associations with gene expression regulation in whole blood -- a tissue commonly used in eQTL studies. Thus, large-scale maps of variants associated with gene regulation lack a condition important for understanding autoimmunity. As a proof-of-principle we identified variant rs6927172, which links stimulated T cell-specific chromatin dysregulation in the TNFAIP3 locus to ulcerative colitis and rheumatoid arthritis. Overall, our results provide a broad resource of chromatin landscape dynamics and highlight the need for large-scale characterization of effects of genetic variation in stimulated cells.BibTeX:
@article{Calderon2018immune, author = {Calderon, Diego and Nguyen, Michelle L.T. and Mezger, Anja and Kathiria, Arwa and Nguyen, Vinh and Lescano, Ninnia and Wu, Beijing and Trombetta, John and Ribado, Jessica V. and Knowles, David A. and Gao, Ziyue and Parent, Audrey V. and Burt, Trevor D. and Anderson, Mark S. and Criswell, Lindsey A. and Greenleaf, William J. and Marson, Alexander and Pritchard, Jonathan K.}, title = {Landscape of stimulation-responsive chromatin across diverse human immune cells}, journal = {Nature Genetics}, year = {2019}, url = {https://www.biorxiv.org/content/early/2018/09/05/409722} } - Knowles* DA, Burrows* CK, Blischak JD, Patterson KM, Serie DJ, Norton N, Ober C, Pritchard JK and Gilad Y (2018), "Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes", eLife. *These authors contributed equally to this work.Abstract: Anthracycline-induced cardiotoxicity (ACT) is a key limiting factor in setting optimal chemotherapy regimes for cancer patients, with almost half of patients expected to ultimately develop congestive heart failure given high drug doses. However, the genetic basis of sensitivity to anthracyclines such as doxorubicin remains unclear. To begin addressing this, we created a panel of iPSC-derived cardiomyocytes from 45 individuals and performed RNA-seq after 24h exposure to varying levels of doxorubicin. The transcriptomic response to doxorubicin is substantial, with the majority of genes being differentially expressed across treatments of different concentrations and over 6000 genes showing evidence of differential splicing. Overall, our observations indicate that splicing fidelity decreases in the presence of doxorubicin. We detect 376 response-expression QTLs and 42 response-splicing QTLs, i.e. genetic variants that modulate the individual transcriptomic response to doxorubicin in terms of expression and splicing changes respectively. We show that inter-individual variation in transcriptional response is predictive of cell damage measured in vitro using a cardiac troponin assay, which in turn is shown to be associated with in vivo ACT risk. Finally, the molecular QTLs we detected are enriched in lower ACT GWAS p-values, further supporting the in vivo relevance of our map of genetic regulation of cellular response to anthracyclines.BibTeX:
@article{Knowles2018dox, author = {Knowles*, David A and Burrows*, Courtney K and Blischak, John D and Patterson, Kristen M and Serie, Daniel J. and Norton, Nadine and Ober, Carole and Pritchard, Jonathan K and Gilad, Yoav}, title = {Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes}, journal = {eLife}, year = {2018}, url = {https://elifesciences.org/articles/33480}, doi = {10.7554/eLife.33480} } - Leland Taylor D, Knowles DA, Scott LJ, Ramirez AH, Casale FP, Wolford BN, Guan L, Varshney A, Albanus RD, Parker SCJ, Narisu N, Chines PS, Erdos MR, Welch RP, Kinnunen L, Saramies J, Sundvall J, Lakka TA, Laakso M, Tuomilehto J, Koistinen HA, Stegle O, Boehnke M, Birney E and Collins FS (2018), "Interactions between genetic variation and cellular environment in skeletal muscle gene expression", PLoS One. Vol. 13(4), pp. e0195788.Abstract: From whole organisms to individual cells, responses to environmental conditions are influenced by genetic makeup, where the effect of genetic variation on a trait depends on the environmental context. RNA-sequencing quantifies gene expression as a molecular trait, and is capable of capturing both genetic and environmental effects. In this study, we explore opportunities of using allele-specific expression (ASE) to discover cis-acting genotype-environment interactions (GxE)---genetic effects on gene expression that depend on an environmental condition. Treating 17 common, clinical traits as approximations of the cellular environment of 267 skeletal muscle biopsies, we identify 10 candidate environmental response expression quantitative trait loci (reQTLs) across 6 traits (12 unique gene-environment trait pairs; 10% FDR per trait) including sex, systolic blood pressure, and low-density lipoprotein cholesterol. Although using ASE is in principle a promising approach to detect GxE effects, replication of such signals can be challenging as validation requires harmonization of environmental traits across cohorts and a sufficient sampling of heterozygotes for a transcribed SNP. Comprehensive discovery and replication will require large human transcriptome datasets, or the integration of multiple transcribed SNPs, coupled with standardized clinical phenotyping.BibTeX:
@article{Leland_Taylor2018-lb, author = {Leland Taylor, D and Knowles, David A and Scott, Laura J and Ramirez, Andrea H and Casale, Francesco Paolo and Wolford, Brooke N and Guan, Li and Varshney, Arushi and Albanus, Ricardo D'oliveira and Parker, Stephen C J and Narisu, Narisu and Chines, Peter S and Erdos, Michael R and Welch, Ryan P and Kinnunen, Leena and Saramies, Jouko and Sundvall, Jouko and Lakka, Timo A and Laakso, Markku and Tuomilehto, Jaakko and Koistinen, Heikki A and Stegle, Oliver and Boehnke, Michael and Birney, Ewan and Collins, Francis S}, title = {Interactions between genetic variation and cellular environment in skeletal muscle gene expression}, journal = {PLoS One}, publisher = {Public Library of Science}, year = {2018}, volume = {13}, number = {4}, pages = {e0195788}, doi = {10.1371/journal.pone.0195788} } - Knowles DA, Bouchard G and Plevritis SK (2019), "Sparse discriminative latent characteristics for predicting cancer drug sensitivity from genomic features", PLoS computational biology.[BibTeX]BibTeX:
@article{Knowles2018lacrosse, author = {Knowles, David A and Bouchard, Gina and Plevritis, Sylvia K}, title = {Sparse discriminative latent characteristics for predicting cancer drug sensitivity from genomic features}, journal = {PLoS computational biology}, year = {2019} } - Knowles DA, Davis JR, Edgington H, Raj A, Favé M-J, Zhu X, Potash JB, Weissman MM, Shi J, Levinson D, Awadalla P, Mostafavi S, Montgomery SB and Battle A (2017), "Allele-specific expression reveals interactions between genetic variation and environment", Nature Methods.Abstract: Identifying interactions between genetics and the environment (GxE) remains challenging. We have developed EAGLE, a hierarchical Bayesian model for identifying GxE interactions based on association between environment and allele-specific expression (ASE). Combining RNA-sequencing of whole blood and extensive environmental annotations collected from 922 human individuals, we identified 35 GxE interactions, compared to only four using standard GxE testing. EAGLE provides new opportunities to identify GxE interactions using functional genomic data.BibTeX:
@article{Knowles2017gxe, author = {Knowles, David A and Davis, Joe R and Edgington, Hilary and Raj, Anil and Favé, Marie-Julie and Zhu, Xiaowei and Potash, James B and Weissman, Myrna M and Shi, Jianxin and Levinson, Doug and Awadalla, Philip and Mostafavi, Sara and Montgomery, Stephen B and Battle, Alexis}, title = {Allele-specific expression reveals interactions between genetic variation and environment}, journal = {Nature Methods}, year = {2017}, url = {http://www.nature.com/nmeth/journal/vaop/ncurrent/full/nmeth.4298.html}, doi = {10.1038/nmeth.4298} } - Tung P-Y, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK and Gilad Y (2017), "Batch effects and the effective design of single-cell gene expression studies", Scientific Reports. Vol. 7, pp. 39921.Abstract: Single cell RNA sequencing (scRNA-seq) can be used to characterize variation in gene expression levels at high resolution. However, the sources of experimental noise in scRNA-seq are not yet well understood. We investigated the technical variation associated with sample processing using the single cell Fluidigm C1 platform. To do so, we processed three C1 replicates from three human induced pluripotent stem cell (iPSC) lines. We added unique molecular identifiers (UMIs) to all samples, to account for amplification bias. We found that the major source of variation in the gene expression data was driven by genotype, but we also observed substantial variation between the technical replicates. We observed that the conversion of reads to molecules using the UMIs was impacted by both biological and technical variation, indicating that UMI counts are not an unbiased estimator of gene expression levels. Based on our results, we suggest a framework for effective scRNA-seq studies.BibTeX:
@article{Tung2017, author = {Tung, Po-Yuan and Blischak, John D. and Hsiao, Chiaowen Joyce and Knowles, David A. and Burnett, Jonathan E. and Pritchard, Jonathan K. and Gilad, Yoav}, title = {Batch effects and the effective design of single-cell gene expression studies}, journal = {Scientific Reports}, year = {2017}, volume = {7}, pages = {39921}, url = {http://www.nature.com/articles/srep39921}, doi = {10.1038/srep39921} } - Calderon D, Bhaskar A, Knowles DA, Golan D, Raj T, Fu AQ and Pritchard JK (2017), "Inferring Relevant Cell Types for Complex Traits by Using Single-Cell Gene Expression", American Journal of Human Genetics.Abstract: Previous studies have prioritized trait-relevant cell types by looking for an enrichment of GWAS signal within functional regions. However, these studies are limited in cell resolution by the lack of functional annotations from difficult-to-characterize or rare cell populations. Measurement of single-cell gene expression has become a popular method for characterizing novel cell types, and yet, hardly any work exists linking single-cell RNA-seq to phenotypes of interest. To address this deficiency, we present RolyPoly, a regression-based polygenic model that can prioritize trait-relevant cell types and genes from GWAS summary statistics and single-cell RNA-seq. We demonstrate RolyPoly's accuracy through simulation and validate previously known tissue-trait associations. We discover a significant association between microglia and late-onset Alzheimer's disease, and an association between oligodendrocytes and replicating fetal cortical cells with schizophrenia. Additionally, RolyPoly computes a trait-relevance score for each gene which reflects the importance of expression specific to a cell type. We found that differentially expressed genes in the prefrontal cortex of Alzheimer's patients were significantly enriched for highly ranked genes by RolyPoly gene scores. Overall, our method represents a powerful framework for understanding the effect of common variants on cell types contributing to complex traits.BibTeX:
@article{Calderon2017, author = {Calderon, Diego and Bhaskar, Anand and Knowles, David A and Golan, David and Raj, Towfique and Fu, Audrey Q and Pritchard, Jonathan K}, title = {Inferring Relevant Cell Types for Complex Traits by Using Single-Cell Gene Expression}, journal = {American Journal of Human Genetics}, year = {2017}, url = {http://www.cell.com/ajhg/abstract/S0002-9297(17)30378-6} } - Tsang EK, Abell NS, Li X, Anaya V, Karczewski KJ, Knowles DA, Sierra RG, Smith KS and Montgomery SB (2017), "Small RNA sequencing in cells and exosomes identifies eQTLs and 14q32 as a region of active export", G3 Genes|Genomes|Genetics. Vol. 7(1), pp. 31-39.Abstract: Exosomes are small extracellular vesicles that carry heterogeneous cargo, including RNA, between cells. Increasing evidence suggests that exosomes are important mediators of intercellular communication and biomarkers of disease. Despite this, the variability of exosomal RNA between individuals has not been well quantified. To assess this variability, we sequenced the small RNA of cells and exosomes from a 17-member family. Across individuals, we show that selective export of miRNAs occurs not only at the level of specific transcripts, but that a cluster of 74 mature miRNAs on chromosome 14q32 is massively exported in exosomes while mostly absent from cells. We also observe more interindividual variability between exosomal samples than between cellular ones and identify four miRNA expression quantitative trait loci shared between cells and exosomes. Our findings indicate that genomically colocated miRNAs can be exported together and highlight the variability in exosomal miRNA levels between individuals as relevant for exosome use as diagnostics.BibTeX:
@article{Tsang2017, author = {Tsang, Emily K. and Abell, Nathan S. and Li, Xin and Anaya, Vanessa and Karczewski, Konrad J. and Knowles, David A. and Sierra, Raymond G. and Smith, Kevin S. and Montgomery, Stephen B.}, title = {Small RNA sequencing in cells and exosomes identifies eQTLs and 14q32 as a region of active export}, journal = {G3 Genes|Genomes|Genetics}, year = {2017}, volume = {7}, number = {1}, pages = {31--39}, url = {http://g3journal.org/lookup/doi/10.1534/g3.116.036137}, doi = {10.1534/g3.116.036137} } - Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, Pulst SM, Taylor JP, Rigo F and Gitler AD (2017), "Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice", Nature. Vol. 544(7650), pp. 367-371.Abstract: Amyotrophic lateral sclerosis (ALS) is a rapidly progressing neurodegenerative disease that is characterized by motor neuron loss and that leads to paralysis and death 2--5 years after disease onset 1 . Nearly all patients with ALS have aggregates of the RNA-binding protein TDP-43 in their brains and spinal cords 2 , and rare mutations in the gene encoding TDP-43 can cause ALS 3 . There are no effective TDP-43-directed therapies for ALS or related TDP-43 proteinopathies, such as frontotemporal dementia. Antisense oligonucleotides (ASOs) and RNA-interference approaches are emerging as attractive therapeutic strategies in neurological diseases 4 . Indeed, treatment of a rat model of inherited ALS (caused by a mutation in Sod1) with ASOs against Sod1 has been shown to substantially slow disease progression 5 . However, as SOD1 mutations account for only around 2--5% of ALS cases, additional therapeutic strategies are needed. Silencing TDP-43 itself is probably not appropriate, given its critical cellular functions 1,6 . Here we present a promising alternative therapeutic strategy for ALS that involves targeting ataxin-2. A decrease in ataxin-2 suppresses TDP-43 toxicity in yeast and flies 7 , and intermediate-length polyglutamine expansions in the ataxin-2 gene increase risk of ALS 7,8 . We used two independent approaches to test whether decreasing ataxin-2 levels could mitigate disease in a mouse model of TDP-43 proteinopathy 9 . First, we crossed ataxin-2 knockout mice with TDP-43 (also known as TARDBP) transgenic mice. The decrease in ataxin-2 reduced aggregation of TDP-43, markedly increased survival and improved motor function. Second, in a more therapeutically applicable approach, we administered ASOs targeting ataxin-2 to the central nervous system of TDP-43 transgenic mice. This single treatment markedly extended survival. Because TDP-43 aggregation is a component of nearly all cases of ALS 6 , targeting ataxin-2 could represent a broadly effective therapeutic strategy. To test the hypothesis that a decrease in ataxin-2 levels can res-cue neurodegenerative phenotypes caused by TDP-43 accumula-tion, we first used a genetic approach. There are several transgenic mouse lines that express wild-type or mutant TDP-43, using various strategies 10BibTeX:
@article{Becker2017, author = {Becker, Lindsay A. and Huang, Brenda and Bieri, Gregor and Ma, Rosanna and Knowles, David A. and Jafar-Nejad, Paymaan and Messing, James and Kim, Hong Joo and Soriano, Armand and Auburger, Georg and Pulst, Stefan M. and Taylor, J. Paul and Rigo, Frank and Gitler, Aaron D.}, title = {Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice}, journal = {Nature}, year = {2017}, volume = {544}, number = {7650}, pages = {367--371}, url = {http://www.nature.com/doifinder/10.1038/nature22038}, doi = {10.1038/nature22038} } - Davis JR, Fresard L, Knowles DA, Pala M, Bustamante CD, Battle A and Montgomery SB (2016), "An Efficient Multiple-Testing Adjustment for eQTL Studies that Accounts for Linkage Disequilibrium between Variants", The American Journal of Human Genetics. Vol. 98(1), pp. 216-224.Abstract: Methods for multiple-testing correction in local expression quantitative trait locus (cis-eQTL) studies are a trade-off between statistical power and computational efficiency. Bonferroni correction, though computationally trivial, is overly conservative and fails to account for linkage disequilibrium between variants. Permutation-based methods are more powerful, though computationally far more intensive. We present an alternative correction method called eigenMT, which runs over 500 times faster than permutations and has adjusted p values that closely approximate empirical ones. To achieve this speed while also maintaining the accuracy of permutation-based methods, we estimate the effective number of independent variants tested for association with a particular gene, termed Meff, by using the eigenvalue decomposition of the genotype correlation matrix. We employ a regularized estimator of the correlation matrix to ensure Meff is robust and yields adjusted p values that closely approximate p values from permutations. Finally, using a common genotype matrix, we show that eigenMT can be applied with even greater efficiency to studies across tissues or conditions. Our method provides a simpler, more efficient approach to multiple-testing correction than existing methods and fits within existing pipelines for eQTL discovery.BibTeX:
@article{Davis2016eigenmt, author = {Davis, Joe R. and Fresard, Laure and Knowles, David A. and Pala, Mauro and Bustamante, Carlos D. and Battle, Alexis and Montgomery, Stephen B.}, title = {An Efficient Multiple-Testing Adjustment for eQTL Studies that Accounts for Linkage Disequilibrium between Variants}, journal = {The American Journal of Human Genetics}, year = {2016}, volume = {98}, number = {1}, pages = {216--224}, url = {http://www.cell.com/ajhg/abstract/S0002-9297(15)00492-9}, doi = {10.1016/j.ajhg.2015.11.021} } - Kukurba KR, Parsana P, Balliu B, Smith KS, Zappala Z, Knowles DA, Favé M-J, Davis JR, Li X, Zhu X, Potash JB, Weissman MM, Shi J, Kundaje A, Levinson DF, Awadalla P, Mostafavi S, Battle A and Montgomery SB (2016), "Impact of the X chromosome and sex on regulatory variation", Genome Research. Vol. 26(6), pp. 768-777.Abstract: The X chromosome, with its unique mode of inheritance, contributes to differences between the sexes at a molecular level, including sex-specific gene expression and sex-specific impact of genetic variation. We have conducted an analysis of the impact of both sex and the X chromosome on patterns of gene expression identified through transcriptome sequencing of whole blood from 922 individuals. We identified that genes on the X chromosome are more likely to have sex-specific expression compared to the autosomal genes. Furthermore, we identified a depletion of regulatory variants on the X chromosome, especially among genes under high selective constraint. In contrast, we discovered an enrichment of sex-specific regulatory variants on the X chromosome. To resolve the molecular mechanisms underlying such effects, we generated and connected sex-specific chromatin accessibility to sex-specific expression and regulatory variation. As sex-specific regulatory variants can inform sex differences in genetic disease prevalence, we have integrated our data with genome-wide association study data for multiple immune traits and to identify traits with significant sex biases. Together, our study provides genome-wide insight into how the X chromosome and sex shape human gene regulation and disease.BibTeX:
@article{Kukurba2015, author = {Kukurba, Kimberly R. and Parsana, Princy and Balliu, Brunilda and Smith, Kevin S. and Zappala, Zachary and Knowles, David A. and Favé, Marie-Julie and Davis, Joe R. and Li, Xin and Zhu, Xiaowei and Potash, James B. and Weissman, Myrna M. and Shi, Jianxin and Kundaje, Anshul and Levinson, Douglas F. and Awadalla, Philip and Mostafavi, Sara and Battle, Alexis and Montgomery, Stephen B.}, title = {Impact of the X chromosome and sex on regulatory variation}, journal = {Genome Research}, publisher = {Cold Spring Harbor Labs Journals}, year = {2016}, volume = {26}, number = {6}, pages = {768--777}, url = {http://genome.cshlp.org/lookup/doi/10.1101/gr.197897.115}, doi = {10.1101/gr.197897.115} } - Li* YI, Knowles* DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK and Pritchard JK (2017), "Annotation-free quantification of RNA splicing using LeafCutter", Nature Genetics. *These authors contributed equally to this work.Abstract: The excision of introns from pre-mRNA is an essential step in mRNA processing. We developed LeafCutter to study sample and population variation in intron splicing. LeafCutter identifies variable intron splicing events from short-read RNA-seq data and finds alternative splicing events of high complexity. Our approach obviates the need for transcript annotations and overcomes the challenges in estimating relative isoform or exon usage in complex splicing events. LeafCutter can be used both for detecting differential splicing between sample groups, and for mapping splicing quantitative trait loci (sQTLs). Compared to contemporary methods, we find over three times more sQTLs, many of which help us ascribe molecular effects to disease-associated variants. LeafCutter is fast, easy to use, and available at https://github.com/davidaknowles/BibTeX:
@article{LeafCutter, author = {Li*, Yang I. and Knowles*, David A. and Humphrey, Jack and Barbeira, Alvaro N. and Dickinson, Scott P. and Im, Hae Kyung and Pritchard, Jonathan K.}, title = {Annotation-free quantification of RNA splicing using LeafCutter}, journal = {Nature Genetics}, year = {2017}, url = {https://www.nature.com/articles/s41588-017-0004-9}, doi = {10.1038/s41588-017-0004-9} } - Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y and Pritchard JK (2016), "RNA splicing is a primary link between genetic variation and disease.", Science. Vol. 352(6285), pp. 600-4.Abstract: Noncoding variants play a central role in the genetics of complex traits, but we still lack a full understanding of the molecular pathways through which they act. We quantified the contribution of cis-acting genetic effects at all major stages of gene regulation from chromatin to proteins, in Yoruba lymphoblastoid cell lines (LCLs). About 65% of expression quantitative trait loci (eQTLs) have primary effects on chromatin, whereas the remaining eQTLs are enriched in transcribed regions. Using a novel method, we also detected 2893 splicing QTLs, most of which have little or no effect on gene-level expression. These splicing QTLs are major contributors to complex traits, roughly on a par with variants that affect gene expression levels. Our study provides a comprehensive view of the mechanisms linking genetic variation to variation in human gene regulation.BibTeX:
@article{Li2016splicing, author = {Li, Yang I and van de Geijn, Bryce and Raj, Anil and Knowles, David A and Petti, Allegra A and Golan, David and Gilad, Yoav and Pritchard, Jonathan K}, title = {RNA splicing is a primary link between genetic variation and disease.}, journal = {Science}, publisher = {American Association for the Advancement of Science}, year = {2016}, volume = {352}, number = {6285}, pages = {600--4}, url = {http://www.ncbi.nlm.nih.gov/pubmed/27126046}, doi = {10.1126/science.aad9417} } - Kukurba KR, Zhang R, Li X, Smith KS, Knowles DA, How Tan M, Piskol R, Lek M, Snyder M, MacArthur DG, Li JB and Montgomery SB (2014), "Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues", PLoS Genetics. Vol. 10(5), pp. e1004304.Abstract: Personal exome and genome sequencing provides access to loss-of-function and rare deleterious alleles whose interpretation is expected to provide insight into individual disease burden. However, for each allele, accurate interpretation of its effect will depend on both its penetrance and the trait's expressivity. In this regard, an important factor that can modify the effect of a pathogenic coding allele is its level of expression; a factor which itself characteristically changes across tissues. To better inform the degree to which pathogenic alleles can be modified by expression level across multiple tissues, we have conducted exome, RNA and deep, targeted allele-specific expression (ASE) sequencing in ten tissues obtained from a single individual. By combining such data, we report the impact of rare and common loss-of-function variants on allelic expression exposing stronger allelic bias for rare stop-gain variants and informing the extent to which rare deleterious coding alleles are consistently expressed across tissues. This study demonstrates the potential importance of transcriptome data to the interpretation of pathogenic protein-coding variants.BibTeX:
@article{Kukurba2014deleterious, author = {Kukurba, Kimberly R. and Zhang, Rui and Li, Xin and Smith, Kevin S. and Knowles, David A. and How Tan, Meng and Piskol, Robert and Lek, Monkol and Snyder, Michael and MacArthur, Daniel G. and Li, Jin Billy and Montgomery, Stephen B.}, title = {Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues}, journal = {PLoS Genetics}, publisher = {Public Library of Science}, year = {2014}, volume = {10}, number = {5}, pages = {e1004304}, url = {http://dx.plos.org/10.1371/journal.pgen.1004304}, doi = {10.1371/journal.pgen.1004304} } - Li X, Battle A, Karczewski KJ, Zappala Z, Knowles DA, Smith KS, Kukurba KR, Wu E, Simon N and Montgomery SB (2014), "Transcriptome sequencing of a large human family identifies the impact of rare noncoding variants.", American Journal of Human Genetics. Vol. 95(3), pp. 245-56.Abstract: Recent and rapid human population growth has led to an excess of rare genetic variants that are expected to contribute to an individual's genetic burden of disease risk. To date, much of the focus has been on rare protein-coding variants, for which potential impact can be estimated from the genetic code, but determining the impact of rare noncoding variants has been more challenging. To improve our understanding of such variants, we combined high-quality genome sequencing and RNA sequencing data from a 17-individual, three-generation family to contrast expression quantitative trait loci (eQTLs) and splicing quantitative trait loci (sQTLs) within this family to eQTLs and sQTLs within a population sample. Using this design, we found that eQTLs and sQTLs with large effects in the family were enriched with rare regulatory and splicing variants (minor allele frequency 0.01). They were also more likely to influence essential genes and genes involved in complex disease. In addition, we tested the capacity of diverse noncoding annotation to predict the impact of rare noncoding variants. We found that distance to the transcription start site, evolutionary constraint, and epigenetic annotation were considerably more informative for predicting the impact of rare variants than for predicting the impact of common variants. These results highlight that rare noncoding variants are important contributors to individual gene-expression profiles and further demonstrate a significant capability for genomic annotation to predict the impact of rare noncoding variants..BibTeX:
@article{Li2014rare, author = {Li, Xin and Battle, Alexis and Karczewski, Konrad J. and Zappala, Zach and Knowles, David A. and Smith, Kevin S. and Kukurba, Kim R. and Wu, Eric and Simon, Noah and Montgomery, Stephen B.}, title = {Transcriptome sequencing of a large human family identifies the impact of rare noncoding variants.}, journal = {American Journal of Human Genetics}, publisher = {Elsevier}, year = {2014}, volume = {95}, number = {3}, pages = {245--56}, url = {http://www.cell.com/article/S0002929714003486/fulltext}, doi = {10.1016/j.ajhg.2014.08.004} } - Glass D, Viñuela A, Davies MN, Ramasamy A, Parts L, Knowles DA, Brown AA, Hedman AK, Small KS, Buil A, Grundberg E, Nica AC, Meglio P, Nestle FO, Ryten M, Durbin R, McCarthy MI, Deloukas P, Dermitzakis ET, Weale ME, Bataille V and Spector TD (2013), "Gene expression changes with age in skin, adipose tissue, blood and brain.", Genome biology. Vol. 14(7), pp. R75.Abstract: BACKGROUND: Previous studies have demonstrated that gene expression levels change with age. These changes are hypothesized to influence the aging rate of an individual. We analyzed gene expression changes with age in abdominal skin, subcutaneous adipose tissue and lymphoblastoid cell lines in 856 female twins in the age range of 39-85 years. Additionally, we investigated genotypic variants involved in genotype-by-age interactions to understand how the genomic regulation of gene expression alters with age. RESULTS: Using a linear mixed model, differential expression with age was identified in 1,672 genes in skin and 188 genes in adipose tissue. Only two genes expressed in lymphoblastoid cell lines showed significant changes with age. Genes significantly regulated by age were compared with expression profiles in 10 brain regions from 100 postmortem brains aged 16 to 83 years. We identified only one age-related gene common to the three tissues. There were 12 genes that showed differential expression with age in both skin and brain tissue and three common to adipose and brain tissues. CONCLUSIONS: Skin showed the most age-related gene expression changes of all the tissues investigated, with many of the genes being previously implicated in fatty acid metabolism, mitochondrial activity, cancer and splicing. A significant proportion of age-related changes in gene expression appear to be tissue-specific with only a few genes sharing an age effect in expression across tissues. More research is needed to improve our understanding of the genetic influences on aging and the relationship with age-related diseases.BibTeX:
@article{Glass2013muther, author = {Glass, Daniel and Viñuela, Ana and Davies, Matthew N and Ramasamy, Adaikalavan and Parts, Leopold and Knowles, David A. and Brown, Andrew A and Hedman, Asa K and Small, Kerrin S and Buil, Alfonso and Grundberg, Elin and Nica, Alexandra C and Meglio, Paoladi and Nestle, Frank O and Ryten, Mina and Durbin, Richard and McCarthy, Mark I and Deloukas, Panagiotis and Dermitzakis, Emmanouil T and Weale, Michael E and Bataille, Veronique and Spector, Tim D}, title = {Gene expression changes with age in skin, adipose tissue, blood and brain.}, journal = {Genome biology}, year = {2013}, volume = {14}, number = {7}, pages = {R75}, url = {http://genomebiology.com/2013/14/7/R75}, doi = {10.1186/gb-2013-14-7-r75} } - Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang T-P, Meduri E, Barrett A, Nisbett J, Sekowska M, Wilk A, Shin S-Y, Glass D, Travers M, Min JL, Knowles DA, Ring S, Ho K, Thorleifsson G, Kong A, Thorsteindottir U, Ainali C, Dimas AS, Hassanali N, Ingle C, Krestyaninova M, Lowe CE, Di Meglio P, Montgomery SB, Parts L, Potter S, Surdulescu G, Tsaprouni L, Tsoka S, Bataille V, Durbin R, Nestle FO, O'Rahilly S, Soranzo N, Lindgren CM, Zondervan KT, Ahmadi KR, Schadt EE, Stefansson K, Smith GD, McCarthy MI, Deloukas P, Dermitzakis ET and Spector TD (2012), "Mapping cis- and trans-regulatory effects across multiple tissues in twins.", Nature Genetics. Vol. 44(10), pp. 1084-9.Abstract: Sequence-based variation in gene expression is a key driver of disease risk. Common variants regulating expression in cis have been mapped in many expression quantitative trait locus (eQTL) studies, typically in single tissues from unrelated individuals. Here, we present a comprehensive analysis of gene expression across multiple tissues conducted in a large set of mono- and dizygotic twins that allows systematic dissection of genetic (cis and trans) and non-genetic effects on gene expression. Using identity-by-descent estimates, we show that at least 40% of the total heritable cis effect on expression cannot be accounted for by common cis variants, a finding that reveals the contribution of low-frequency and rare regulatory variants with respect to both transcriptional regulation and complex trait susceptibility. We show that a substantial proportion of gene expression heritability is trans to the structural gene, and we identify several replicating trans variants that act predominantly in a tissue-restricted manner and may regulate the transcription of many genes.BibTeX:
@article{Grundberg2012, author = {Grundberg, Elin and Small, Kerrin S and Hedman, Asa K and Nica, Alexandra C and Buil, Alfonso and Keildson, Sarah and Bell, Jordana T and Yang, Tsun-Po and Meduri, Eshwar and Barrett, Amy and Nisbett, James and Sekowska, Magdalena and Wilk, Alicja and Shin, So-Youn and Glass, Daniel and Travers, Mary and Min, Josine L and Knowles, David A. and Ring, Sue and Ho, Karen and Thorleifsson, Gudmar and Kong, Augustine and Thorsteindottir, Unnur and Ainali, Chrysanthi and Dimas, Antigone S and Hassanali, Neelam and Ingle, Catherine and Krestyaninova, Maria and Lowe, Christopher E and Di Meglio, Paola and Montgomery, Stephen B and Parts, Leopold and Potter, Simon and Surdulescu, Gabriela and Tsaprouni, Loukia and Tsoka, Sophia and Bataille, Veronique and Durbin, Richard and Nestle, Frank O and O'Rahilly, Stephen and Soranzo, Nicole and Lindgren, Cecilia M and Zondervan, Krina T and Ahmadi, Kourosh R and Schadt, Eric E and Stefansson, Kari and Smith, George Davey and McCarthy, Mark I and Deloukas, Panos and Dermitzakis, Emmanouil T and Spector, Tim D.}, title = {Mapping cis- and trans-regulatory effects across multiple tissues in twins.}, journal = {Nature Genetics}, year = {2012}, volume = {44}, number = {10}, pages = {1084--9}, url = {http://dx.doi.org/10.1038/ng.2394}, doi = {10.1038/ng.2394} } - Schöne C, Venner A, Knowles DA, Karnani MM and Burdakov D (2011), "Dichotomous cellular properties of mouse orexin/hypocretin neurons.", The Journal of Physiology. Vol. 589(Pt 11), pp. 2767-79.Abstract: Hypothalamic hypocretin/orexin (Hcrt/Orx) neurons recently emerged as critical regulators of sleep--wake cycles, reward seeking and body energy balance. However, at the level of cellular and network properties, it remains unclear whether Hcrt/Orx neurons are one homogeneous population, or whether there are several distinct types of Hcrt/Orx cells. Here, we collated diverse structural and functional information about individual Hcrt/Orx neurons in mouse brain slices, by combining patch-clamp analysis of spike firing, membrane currents and synaptic inputs with confocal imaging of cell shape and subsequent 3-dimensional Sholl analysis of dendritic architecture. Statistical cluster analysis of intrinsic firing properties revealed that Hcrt/Orx neurons fall into two distinct types. These two cell types also differ in the complexity of their dendritic arbour, the strength of AMPA and GABAA receptor-mediated synaptic drive that they receive, and the density of low-threshold, 4-aminopyridine-sensitive, transient K+ current. Our results provide quantitative evidence that, at the cellular level, the mouse Hcrt/Orx system is composed of two classes of neurons with different firing properties, morphologies and synaptic input organization.BibTeX:
@article{Schone2011, author = {Schöne, Cornelia and Venner, Anne and Knowles, David A. and Karnani, Mahesh M and Burdakov, Denis}, title = {Dichotomous cellular properties of mouse orexin/hypocretin neurons.}, journal = {The Journal of Physiology}, year = {2011}, volume = {589}, number = {Pt 11}, pages = {2767--79}, url = {http://jp.physoc.org/content/early/2011/04/11/jphysiol.2011.208637.abstract}, doi = {10.1113/jphysiol.2011.208637} } - Movassagh M, Choy M-K, Knowles DA, Cordeddu L, Haider S, Down T, Siggens L, Vujic A, Simeoni I, Penkett C, Goddard M, Lio P, Bennett M and Foo R (2011), "Distinct Epigenomic Features in End-Stage Failing Human Hearts", Circulation, American Heart Association. Vol. 135Abstract: BACKGROUND: The epigenome refers to marks on the genome, including DNA methylation and histone modifications, that regulate the expression of underlying genes. A consistent profile of gene expression changes in end-stage cardiomyopathy led us to hypothesize that distinct global patterns of the epigenome may also exist. METHODS AND RESULTS: We constructed genome-wide maps of DNA methylation and histone-3 lysine-36 trimethylation (H3K36me3) enrichment for cardiomyopathic and normal human hearts. More than 506 Mb sequences per library were generated by high-throughput sequencing, allowing us to assign methylation scores to ≈28 million CG dinucleotides in the human genome. DNA methylation was significantly different in promoter CpG islands, intragenic CpG islands, gene bodies, and H3K36me3-enriched regions of the genome. DNA methylation differences were present in promoters of upregulated genes but not downregulated genes. H3K36me3 enrichment itself was also significantly different in coding regions of the genome. Specifically, abundance of RNA transcripts encoded by the DUX4 locus correlated to differential DNA methylation and H3K36me3 enrichment. In vitro, Dux gene expression was responsive to a specific inhibitor of DNA methyltransferase, and Dux siRNA knockdown led to reduced cell viability. CONCLUSIONS: Distinct epigenomic patterns exist in important DNA elements of the cardiac genome in human end-stage cardiomyopathy. The epigenome may control the expression of local or distal genes with critical functions in myocardial stress response. If epigenomic patterns track with disease progression, assays for the epigenome may be useful for assessing prognosis in heart failure. Further studies are needed to determine whether and how the epigenome contributes to the development of cardiomyopathy.BibTeX:
@article{Movassagh2011a, author = {Movassagh, Mehregan and Choy, Mun-Kit and Knowles, David A and Cordeddu, Lina and Haider, Syed and Down, Thomas and Siggens, Lee and Vujic, Ana and Simeoni, Ilenia and Penkett, Chris and Goddard, Martin and Lio, Pietro and Bennett, Martin and Foo, Roger}, title = {Distinct Epigenomic Features in End-Stage Failing Human Hearts}, journal = {Circulation, American Heart Association}, year = {2011}, volume = {135}, url = {http://circ.ahajournals.org/content/early/2011/10/24/CIRCULATIONAHA.111.040071.abstract}, doi = {10.1161/CIRCULATIONAHA.111.040071} } - Glass D, Parts L, Knowles D, Aviv A and Spector TD (2010), "No correlation between childhood maltreatment and telomere length.", Biological psychiatry. Vol. 68(6), pp. e21-2.BibTeX:
@article{Glass2010, author = {Glass, Daniel and Parts, Leopold and Knowles, David and Aviv, Abraham and Spector, Tim D}, title = {No correlation between childhood maltreatment and telomere length.}, journal = {Biological psychiatry}, year = {2010}, volume = {68}, number = {6}, pages = {e21--2}, url = {http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2930212/}, doi = {10.1016/j.biopsych.2010.02.026} }
Machine learning/statistics
- Stirn A, Wessels H-H, Schertzer M, Pereira L, Sanjana NE and Knowles DA (2023), "Faithful Heteroscedastic Regression with Neural Networks", In 26th International Conference on Artificial Intelligence and Statistics (AISTATS).Abstract: Heteroscedastic regression models a Gaussian variable's mean and variance as a function of covariates. Parametric methods that employ neural networks for these parameter maps can capture complex relationships in the data. Yet, optimizing network parameters via log likelihood gradients can yield suboptimal mean and uncalibrated variance estimates. Current solutions side-step this optimization problem with surrogate objectives or Bayesian treatments. Instead, we make two simple modifications to optimization. Notably, their combination produces a heteroscedastic model with mean estimates that are provably as accurate as those from its homoscedastic counterpart (i.e. fitting the mean under squared error loss). For a wide variety of network and task complexities, we find that mean estimates from existing heteroscedastic solutions can be significantly less accurate than those from an equivalently expressive mean-only model. Our approach provably retains the accuracy of an equally flexible mean-only model while also offering best-in-class variance calibration. Lastly, we show how to leverage our method to recover the underlying heteroscedastic noise variance.BibTeX:
@inproceedings{Stirn2023, author = {Stirn, Andrew and Wessels, Hans-Hermann and Schertzer, Megan and Pereira, Laura and Sanjana, Neville E and Knowles, David A}, title = {Faithful Heteroscedastic Regression with Neural Networks}, booktitle = {26th International Conference on Artificial Intelligence and Statistics (AISTATS)}, year = {2023}, url = {http://arxiv.org/abs/2212.09184} } - Stirn A, Jebara T and Knowles DA (2019), "A New Distribution on the Simplex with Auto-Encoding Applications", In Advances in Neural Information Processing Systems.Abstract: We construct a new distribution for the simplex using the Kumaraswamy distribution and an ordered stick-breaking process. We explore and develop the theoretical properties of this new distribution and prove that it exhibits symmetry (exchangeability) under the same conditions as the well-known Dirichlet. Like the Dirichlet, the new distribution is adept at capturing sparsity but, unlike the Dirichlet, has an exact and closed form reparameterization--making it well suited for deep variational Bayesian modeling. We demonstrate the distribution's utility in a variety of semi-supervised auto-encoding tasks. In all cases, the resulting models achieve competitive performance commensurate with their simplicity, use of explicit probability models, and abstinence from adversarial training.BibTeX:
@inproceedings{Stirn2019, author = {Andrew Stirn and Tony Jebara and David A. Knowles}, title = {A New Distribution on the Simplex with Auto-Encoding Applications}, booktitle = {Advances in Neural Information Processing Systems}, year = {2019}, url = {http://papers.nips.cc/paper/9520-a-new-distribution-on-the-simplex-with-auto-encoding-applications} } - Palla* K, Knowles* DA and Ghahramani Z (2017), "A birth-death process for feature allocation.", In Proceedings of the 34th International Conference on Machine Learning. *These authors contributed equally to this work.Abstract: We propose a Bayesian nonparametric prior over feature allocations for sequential data, the birth-death feature allocation process (BDFP). The BDFP models the evolution of the feature allocation of a set of N objects across a covariate (e.g.time) by creating and deleting features. A BDFP is exchangeable, projective, stationary and reversible, and its equilibrium distribution is given by the Indian buffet process (IBP). We also show that the Beta process on an extended space is the de Finetti mixing distribution underlying the BDFP. Finally, we present the finite approximation of the BDFP, the Beta Event Process (BEP), that permits simplified inference. The utility of the BDFP as a prior is demonstrated on real world dynamic genomics and social network data.BibTeX:
@inproceedings{palla2017bdfp, author = {Palla*, Konstantina and Knowles*, David A. and Ghahramani, Zoubin}, title = {A birth-death process for feature allocation.}, booktitle = {Proceedings of the 34th International Conference on Machine Learning}, year = {2017} } - Shah A, Knowles DA and Ghahramani Z (2015), "An Empirical Study of Stochastic Variational Inference Algorithms for the Beta Bernoulli Process", In Proceedings of the 32nd International Conference on Machine Learning. , pp. 1594-1603.Abstract: Stochastic variational inference (SVI) is emerging as the most promising candidate for scaling inference in Bayesian probabilistic models to large datasets. However, the performance of these methods has been assessed primarily in the context of Bayesian topic models, particularly latent Dirichlet allocation (LDA). Deriving several new algorithms, and using synthetic, image and genomic datasets, we investigate whether the understanding gleaned from LDA applies in the setting of sparse latent factor models, specifically beta process factor analysis (BPFA). We demonstrate that the big picture is consistent: using Gibbs sampling within SVI to maintain certain posterior dependencies is extremely effective. However, we find that different posterior dependencies are important in BPFA relative to LDA. Particularly, approximations able to model intra-local variable dependence perform best.BibTeX:
@inproceedings{Shah2015, author = {Shah, Amar and Knowles, David A and Ghahramani, Zoubin}, title = {An Empirical Study of Stochastic Variational Inference Algorithms for the Beta Bernoulli Process}, booktitle = {Proceedings of the 32nd International Conference on Machine Learning}, year = {2015}, pages = {1594--1603}, url = {http://proceedings.mlr.press/v37/shahb15.pdf} } - Knowles DA and Ghahramani Z (2015), "Pitman Yor Diffusion Trees for Bayesian hierarchical clustering", IEEE Transactions on Pattern Analysis and Machine Intelligence. Vol. 37(2), pp. 271-289.Abstract: In this paper we introduce the Pitman Yor Diffusion Tree (PYDT), a Bayesian non-parametric prior over tree structures which generalises the Dirichlet Diffusion Tree [Neal, 2001] and removes the restriction to binary branching structure. The generative process is described and shown to result in an exchangeable distribution over data points. We prove some theoretical properties of the model including showing its construction as the continuum limit of a nested Chinese restaurant process model. We then present two alternative MCMC samplers which allows us to model uncertainty over tree structures, and a computationally efficient greedy Bayesian EM search algorithm. Both algorithms use message passing on the tree structure. The utility of the model and algorithms is demonstrated on synthetic and real world data, both continuous and binary.BibTeX:
@article{Knowles2014, author = {Knowles, David A. and Ghahramani, Zoubin}, title = {Pitman Yor Diffusion Trees for Bayesian hierarchical clustering}, journal = {IEEE Transactions on Pattern Analysis and Machine Intelligence}, publisher = {IEEE Computer Society}, year = {2015}, volume = {37}, number = {2}, pages = {271--289}, url = {https://dx.doi.org/10.1109/TPAMI.2014.2313115}, doi = {10.1109/TPAMI.2014.2313115} } - Palla K, Knowles DA and Ghahramani Z (2015), "Relational learning and network modelling using infinite latent attribute models", IEEE Transactions on Pattern Analysis and Machine Intelligence Special Issue on Bayesian Nonparametrics. Vol. 37(2), pp. 462-474.Abstract: Latent variable models for network data extract a summary of the relational structure underlying an observed network. The simplest possible models subdivide nodes of the network into clusters; the probability of a link between any two nodes then depends only on their cluster assignment. Currently available models can be classified by whether clusters are disjoint or are allowed to overlap. These models can explain a flat clustering structure. Hierarchical Bayesian models provide a natural approach to capture more complex dependencies. We propose a model in which objects are characterised by a latent feature vector. Each feature is itself partitioned into disjoint groups (subclusters), corresponding to a second layer of hierarchy. In experimental comparisons, the model achieves significantly improved predictive performance on social and biological link prediction tasks. The results indicate that models with a single layer hierarchy over-simplify real networks.BibTeX:
@article{Palla2015, author = {Palla, Konstantina and Knowles, David A. and Ghahramani, Zoubin}, title = {Relational learning and network modelling using infinite latent attribute models}, journal = {IEEE Transactions on Pattern Analysis and Machine Intelligence Special Issue on Bayesian Nonparametrics}, year = {2015}, volume = {37}, number = {2}, pages = {462--474}, doi = {10.1109/TPAMI.2014.2324586} } - Nguyen K, Bredno J and Knowles DA (2015), "Using contextual information to classify nuclei in histology images", In IEEE 12th International Symposium on Biomedical Imaging (ISBI). , pp. 995-998.Abstract: Nucleus classification is a central task in digital pathology. Given a tissue image, our goal is to classify detected nuclei into different types, for example nuclei of tumor cells, stroma cells, or immune cells. State-of-the-art methods achieve this by extracting different types of features such as morphology, image intensities, and texture features in the nucleus regions. Such features are input to training and classification, e.g. using a support vector machine. In this paper, we introduce additional contextual information obtained from neighboring nuclei or texture in the surrounding tissue regions to improve nucleus classification. Three different methods are presented. These methods use conditional random fields (CRF), texture features computed in image patches centered at each nucleus, and a novel method based on the bag-of-word (BoW) model. The methods are evaluated on images of tumor-burdened tissue from H&E-stained and Ki-67-stained breast samples. The experimental results show that contextual information systematically improves classification accuracy. The proposed BoW-based method performs better than the CRF-based method, and requires less computation than the texture-feature-based method.BibTeX:
@inproceedings{nguyen2015using, author = {Nguyen, Kien and Bredno, Joerg and Knowles, David A}, title = {Using contextual information to classify nuclei in histology images}, booktitle = {IEEE 12th International Symposium on Biomedical Imaging (ISBI)}, year = {2015}, pages = {995--998}, url = {http://dx.doi.org/10.1109/ISBI.2015.7164038} } - Knowles DA, Palla K and Ghahramani Z (2014), "A reversible infinite HMM using normalised random measures", In Proceedings of The 31st International Conference on Machine Learning.Abstract: We present a nonparametric prior over reversible Markov chains. We use completely random measures, specifically gamma processes, to construct a countably infinite graph with weighted edges. By enforcing symmetry to make the edges undirected we define a prior over random walks on graphs that results in a reversible Markov chain. The resulting prior over infinite transition matrices is closely related to the hierarchical Dirichlet process but enforces reversibility. A reinforcement scheme has recently been proposed with similar properties, but the de Finetti measure is not well characterised. We take the alternative approach of explicitly constructing the mixing measure, which allows more straightforward and efficient inference at the cost of no longer having a closed form predictive distribution. We use our process to construct a reversible infinite HMM which we apply to two real datasets, one from epigenomics and one ion channel recording.BibTeX:
@inproceedings{Knowles2014a, author = {Knowles, David A. and Palla, Konstantina and Ghahramani, Zoubin}, title = {A reversible infinite HMM using normalised random measures}, booktitle = {Proceedings of The 31st International Conference on Machine Learning}, year = {2014}, url = {http://proceedings.mlr.press/v32/knowles14.pdf} } - Heaukulani C, Knowles DA and Ghahramani Z (2014), "Beta Diffusion Trees", In Proceedings of the 31st International Conference on Machine Learning. , pp. 1809-1817.Abstract: We define the beta diffusion tree, a random tree structure with a set of leaves that defines a collection of overlapping subsets of objects, known as a feature allocation. A generative process for the tree structure is defined in terms of particles (representing the objects) diffusing in some continuous space, analogously to the Dirichlet diffusion tree (Neal, 2003b), which defines a tree structure over partitions (i.e., non-overlapping subsets) of the objects. Unlike in the Dirichlet diffusion tree, multiple copies of a particle may exist and diffuse along multiple branches in the beta diffusion tree, and an object may therefore belong to multiple subsets of particles. We demonstrate how to build a hierarchically-clustered factor analysis model with the beta diffusion tree and how to perform inference over the random tree structures with a Markov chain Monte Carlo algorithm. We conclude with several numerical experiments on missing data problems with data sets of gene expression microarrays, international development statistics, and intranational socioeconomic measurements.BibTeX:
@inproceedings{Heaukulani2014beta, author = {Heaukulani, Creighton and Knowles, David A. and Ghahramani, Zoubin}, title = {Beta Diffusion Trees}, booktitle = {Proceedings of the 31st International Conference on Machine Learning}, year = {2014}, pages = {1809--1817}, url = {http://proceedings.mlr.press/v32/heaukulani14.pdf} } - Salimans T and Knowles DA (2013), "Fixed-form variational posterior approximation through stochastic linear regression", Bayesian Analysis. Vol. 8(4), pp. 837-882. Winner of the International Society for Bayesian Analysis Lindley Prize..Abstract: We propose a general algorithm for approximating nonstandard Bayesian posterior distributions. The algorithm minimizes the Kullback-Leibler divergence of an approximating distribution to the intractable posterior distribution. Our method can be used to approximate any posterior distribution, provided that it is given in closed form up to the proportionality constant. The approximation can be any distribution in the exponential family or any mixture of such distributions, which means that it can be made arbitrarily precise. Several examples illustrate the speed and accuracy of our approximation method in practice.BibTeX:
@article{salimans2013, author = {Salimans, Tim and Knowles, David A.}, title = {Fixed-form variational posterior approximation through stochastic linear regression}, journal = {Bayesian Analysis}, publisher = {International Society for Bayesian Analysis}, year = {2013}, volume = {8}, number = {4}, pages = {837--882}, url = {http://projecteuclid.org/euclid.ba/1386166315}, doi = {10.1214/13-BA858} } - Quadrianto N, Sharmanska V, Knowles DA and Ghahramani Z (2013), "The Supervised IBP: Neighbourhood Preserving Infinite Latent Feature Models", In Proceedings of the 29th Conference on Uncertainty in Artificial Intelligence.Abstract: We propose a probabilistic model to infer supervised latent variables in the Hamming space from observed data. Our model allows simultaneous inference of the number of binary latent variables, and their values. The latent variables preserve neighbourhood structure of the data in a sense that objects in the same semantic concept have similar latent values, and objects in different concepts have dissimilar latent values. We formulate the supervised infinite latent variable problem based on an intuitive principle of pulling objects together if they are of the same type, and pushing them apart if they are not. We then combine this principle with a flexible Indian Buffet Process prior on the latent variables. We show that the inferred supervised latent variables can be directly used to perform a nearest neighbour search for the purpose of retrieval. We introduce a new application of dynamically extending hash codes, and show how to effectively couple the structure of the hash codes with continuously growing structure of the neighbourhood preserving infinite latent feature space.BibTeX:
@inproceedings{quadrianto2013supervised, author = {Quadrianto, Novi and Sharmanska, Viktoriia and Knowles, David A and Ghahramani, Zoubin}, title = {The Supervised IBP: Neighbourhood Preserving Infinite Latent Feature Models}, booktitle = {Proceedings of the 29th Conference on Uncertainty in Artificial Intelligence}, year = {2013}, url = {http://mlg.eng.cam.ac.uk/pub/pdf/QuaShaKnoGha13.pdf} } - Palla* K, Knowles* DA and Ghahramani Z (2012), "A nonparametric variable clustering model", In Advances in Neural Information Processing Systems. Vol. 5, pp. 2987-2995. *These authors contributed equally to this work.Abstract: Factor analysis models effectively summarise the covariance structure of high dimensional data, but the solutions are typically hard to interpret. This motivates attempting to find a disjoint partition, i.e. a clustering, of observed variables so that variables in a cluster are highly correlated. We introduce a Bayesian non-parametric approach to this problem, and demonstrate advantages over heuristic methods proposed to date.BibTeX:
@inproceedings{Palla2012nonparametric, author = {Palla*, Konstantina and Knowles*, David A. and Ghahramani, Zoubin}, title = {A nonparametric variable clustering model}, booktitle = {Advances in Neural Information Processing Systems}, year = {2012}, volume = {5}, pages = {2987--2995}, url = {https://papers.nips.cc/paper/4579-a-nonparametric-variable-clustering-model} } - Palla* K, Knowles* DA and Ghahramani Z (2012), "An Infinite Latent Attribute Model for Network Data", In Proceedings of the 29th International Conference on Machine Learning. , pp. 1607-1614. *These authors contributed equally to this work.Abstract: Latent variable models for network data extract a summary of the relational structure underlying an observed network. The simplest possible models subdivide nodes of the network into clusters; the probability of a link between any two nodes then depends only on their cluster assignment. Currently available models can be classified by whether clusters are disjoint or are allowed to overlap. These models can explain clustering structure. Hierarchical Bayesian models provide a natural approach to capture more complex dependencies. We propose a model in which objects are characterised by a latent feature vector. Each feature is itself partitioned into disjoint groups (subclusters), corresponding to a second layer of hierarchy. In experimental comparisons, the model achieves significantly improved predictive performance on social and biological link prediction tasks. The results indicate that models with a single layer hierarchy over-simplify real networks.BibTeX:
@inproceedings{palla2012infinite, author = {Palla*, Konstantina and Knowles*, David A. and Ghahramani, Zoubin}, title = {An Infinite Latent Attribute Model for Network Data}, booktitle = {Proceedings of the 29th International Conference on Machine Learning}, year = {2012}, pages = {1607--1614}, url = {http://icml.cc/2012/papers/785.pdf} } - Wilson AG, Knowles DA and Ghahramani Z (2012), "Gaussian Process Regression Networks", In Proceedings of the 29th International Conference on Machine Learning. , pp. 599-606.Abstract: We introduce a new regression frame- work, Gaussian process regression networks (GPRN), which combines the structural properties of Bayesian neural networks with the nonparametric flexibility of Gaussian pro- cesses. GPRN accommodates input (pre- dictor) dependent signal and noise corre- lations between multiple output (response) variables, input dependent length-scales and amplitudes, and heavy-tailed predictive dis- tributions. We derive both elliptical slice sampling and variational Bayes inference pro- cedures for GPRN. We apply GPRN as a multiple output regression and multivariate volatility model, demonstrating substantially improved performance over eight popular multiple output (multi-task) Gaussian pro- cess models and three multivariate volatility models on real datasets, including a 1000 di- mensional gene expression dataset.BibTeX:
@inproceedings{wilson2011gaussian, author = {Wilson, Andrew Gordon and Knowles, David A. and Ghahramani, Zoubin}, title = {Gaussian Process Regression Networks}, booktitle = {Proceedings of the 29th International Conference on Machine Learning}, year = {2012}, pages = {599--606}, url = {http://icml.cc/2012/papers/329.pdf} } - Knowles DA, Gael JV and Ghahramani Z (2011), "Message Passing Algorithms for the Dirichlet Diffusion Tree", In Proceedings of the 28th International Conference on Machine Learning. , pp. 721-728.Abstract: We demonstrate efficient approximate inference for the Dirichlet Diffusion Tree (Neal, 2003), a Bayesian nonparametric prior over tree structures. Although DDTs provide a powerful and elegant approach for modeling hierarchies they haven't seen much use to date. One problem is the computational cost of MCMC inference. We provide the first deterministic approximate inference methods for DDT models and show excellent performance compared to the MCMC alternative. We present message passing algorithms to approximate the Bayesian model evidence for a specific tree. This is used to drive sequential tree building and greedy search to find optimal tree structures, corresponding to hierarchical clusterings of the data. We demonstrate appropriate observation models for continuous and binary data. The empirical performance of our method is very close to the computationally expensive MCMC alternative on a density estimation problem, and significantly outperforms kernel density estimators.BibTeX:
@inproceedings{Gael2011, author = {Knowles, David A. and Gael, Jurgen Van and Ghahramani, Zoubin}, title = {Message Passing Algorithms for the Dirichlet Diffusion Tree}, booktitle = {Proceedings of the 28th International Conference on Machine Learning}, year = {2011}, pages = {721--728}, url = {http://www.icml-2011.org/papers/410_icmlpaper.pdf} } - Knowles DA and Minka T (2011), "Non-conjugate Variational Message Passing for Multinomial and Binary Regression", In Advances in Neural Information Processing Systems. , pp. 1701-1709.Abstract: Variational Message Passing (VMP) is an algorithmic implementation of the Variational Bayes (VB) method which applies only in the special case of conjugate exponential family models. We propose an extension to VMP, which we refer to as Non-conjugate Variational Message Passing (NCVMP) which aims to alleviate this restriction while maintaining modularity, allowing choice in how expectations are calculated, and integrating into an existing message-passing framework: Infer.NET. We demonstrate NCVMP on logistic binary and multinomial regression. In the multinomial case we introduce a novel variational bound for the softmax factor which is tighter than other commonly used bounds whilst maintaining computational tractability.BibTeX:
@inproceedings{Knowles2011c, author = {Knowles, David A. and Minka, Tom}, title = {Non-conjugate Variational Message Passing for Multinomial and Binary Regression}, booktitle = {Advances in Neural Information Processing Systems}, year = {2011}, pages = {1701--1709}, url = {http://papers.nips.cc/paper/4407-non-conjugate-variational-message-passing-for-multinomial-and-binary-regression} } - Knowles DA and Ghahramani Z (2011), "Nonparametric Bayesian sparse factor models with application to gene expression modeling", The Annals of Applied Statistics. Vol. 5(2B), pp. 1534-1552.Abstract: A nonparametric Bayesian extension of Factor Analysis (FA) is proposed where observed data Y is modeled as a linear superposition, G, of a potentially infinite number of hidden factors, X. The Indian Buffet Process (IBP) is used as a prior on G to incorporate sparsity and to allow the number of latent features to be inferred. The model's utility for modeling gene expression data is investigated using randomly generated datasets based on a known sparse connectivity matrix for E. Coli, and on three biological datasets of increasing complexity.BibTeX:
@article{Knowles2011nonparametric, author = {Knowles, David A. and Ghahramani, Zoubin}, title = {Nonparametric Bayesian sparse factor models with application to gene expression modeling}, journal = {The Annals of Applied Statistics}, publisher = {Institute of Mathematical Statistics}, year = {2011}, volume = {5}, number = {2B}, pages = {1534--1552}, url = {https://projecteuclid.org/euclid.aoas/1310562732}, doi = {10.1214/10-AOAS435} } - Knowles DA and Ghahramani Z (2011), "Pitman-Yor Diffusion Trees", In Proceedings of the 27th Conference on Uncertainty in Artificial Intelligence. , pp. 410-418.Abstract: We introduce the Pitman Yor Diffusion Tree (PYDT) for hierarchical clustering, a generalization of the Dirichlet Diffusion Tree (Neal, 2001) which removes the restriction to binary branching structure. The generative process is described and shown to result in an exchangeable distribution over data points. We prove some theoretical properties of the model and then present two inference methods: a collapsed MCMC sampler which allows us to model uncertainty over tree structures, and a computationally efficient greedy Bayesian EM search algorithm. Both algorithms use message passing on the tree structure. The utility of the model and algorithms is demonstrated on synthetic and real world data, both continuous and binary.BibTeX:
@inproceedings{Knowles2011b, author = {Knowles, David A. and Ghahramani, Zoubin}, title = {Pitman-Yor Diffusion Trees}, booktitle = {Proceedings of the 27th Conference on Uncertainty in Artificial Intelligence}, year = {2011}, pages = {410--418}, url = {http://dl.acm.org/citation.cfm?id=3020596} } - Doshi-Velez* F, Mohamed* S, Knowles* DA and Ghahramani Z (2009), "Large Scale Nonparametric Bayesian Inference: Data Parallelisation in the Indian Buffet Process", In Advances in Neural Information Processing Systems. , pp. 1294-1302. *These authors contributed equally to this work.Abstract: Nonparametric Bayesian models provide a framework for flexible probabilistic modelling of complex datasets. Unfortunately, Bayesian inference methods often require high-dimensional averages and can be slow to compute, especially with the potentially unbounded representations associated with nonparametric models. We address the challenge of scaling nonparametric Bayesian inference to the increasingly large datasets found in real-world applications, focusing on the case of parallelising inference in the Indian Buffet Process (IBP). Our approach divides a large data set between multiple processors. The processors use message passing to compute likelihoods in an asynchronous, distributed fashion and to propagate statistics about the global Bayesian posterior. This novel MCMC sampler is the first parallel inference scheme for IBP-based models, scaling to datasets orders of magnitude larger than had previously been possible.BibTeX:
@inproceedings{Doshi-velez2009, author = {Doshi-Velez*, Finale and Mohamed*, Shakir and Knowles*, David A. and Ghahramani, Zoubin}, title = {Large Scale Nonparametric Bayesian Inference: Data Parallelisation in the Indian Buffet Process}, booktitle = {Advances in Neural Information Processing Systems}, year = {2009}, pages = {1294--1302}, url = {http://papers.nips.cc/paper/3669-large-scale-nonparametric-bayesian-inference-data-parallelisation-in-the-indian-buffet-process} } - Knowles DA and Ghahramani Z (2007), "Infinite Sparse Factor Analysis and Infinite Independent Components Analysis", In 7th International Conference on Independent Component Analysis and Signal Separation.Abstract: A nonparametric Bayesian extension of Independent Components Analysis (ICA) is proposed where observed data Y is modelled as a linear superposition, G, of a potentially infinite number of hidden sources, X. Whether a given source is active for a specific data point is specified by an infinite binary matrix, Z. The resulting sparse representation allows increased data reduction compared to standard ICA. We define a prior on Z using the Indian Buffet Process (IBP). We describe four variants of the model, with Gaussian or Laplacian priors on X and the one or two-parameter IBPs. We demonstrate Bayesian inference under these models using a Markov Chain Monte Carlo (MCMC) algorithm on synthetic and gene expression data and compare to standard ICA algorithms.BibTeX:
@inproceedings{Knowles07iica, author = {Knowles, David A. and Ghahramani, Zoubin}, title = {Infinite Sparse Factor Analysis and Infinite Independent Components Analysis}, booktitle = {7th International Conference on Independent Component Analysis and Signal Separation}, year = {2007}, url = {http://www.springerlink.com/index/10.1007/978-3-540-74494-8}, doi = {10.1007/978-3-540-74494-8} }
Software
- Faithful Heteroscedastic Regression with Neural Networks
- Smoother: A Unified Modular Framework to Incorporate Structural Dependency in Spatial Omics Data
- Deep Mendelian randomization: Investigating the causal knowledge of genomic deep learning models
- Phenome-scale causal network discovery with bidirectional mediated Mendelian randomization
- Welch-weighted Egger regression reduces false positives due to correlated pleiotropy in Mendelian randomization bimmer
- Annotation-free quantification of RNA splicing using LeafCutter.
- Detecting gene x environment interaction effects using allele-specific expression..
Reports/Abstracts/Presentations
Workshop papers/conference abstracts
- David A. Knowles, Stanley Ho, Kien Nguyen, Don Morris, Anthony Magliocco, Anindya Sarkar, Daphne Koller, Sylvia Plevritis, Srinivas Chukka, Michael Barnes (2015).
Machine Learning-based Prognostication of Breast Cancer Recurrence using Tissue Slide Features.
Pathology Visions Winner: Best Poster in Image Analysis!
- David A. Knowles, Stanley Ho, Kien Nguyen, Don Morris, Anthony Magliocco, Anindya Sarkar, Daphne Koller, Srinivas Chukka, Michael Barnes (2014)
Machine learning-based prognostication of breast cancer recurrence using tissue slide features from H&E and immunohistochemically stained slides.
San Antonio Breast Cancer Symposium
- David A. Knowles, Leopold Parts, Daniel Glass and John M. Winn
Inferring a measure of physiological age from multiple ageing related phenotypes. paper video
To appear at the NIPS workshop: From Statistical Genetics to Predictive Models in Personalized Medicine (NIPS PM 2011) - David A. Knowles, Leopold Parts, Daniel Glass and John M. Winn (2010)
Modeling skin and ageing phenotypes using latent variable models in Infer.NET. paper poster
Poster presented at: Predictive Models in Personalized Medicine Workshop, NIPS 2010, 6-11 December 2010, Vancouver, BC, Canada. - Knowles, D. and Holmes, S. (2009)
Statistical tools for ultra-deep pyrosequencing of fast evolving viruses. pdf video slides
Presented at: Computational Biology Workshop, NIPS 2009, 7-12 December 2009, Vancouver, BC, Canada.
Reports/Theses
- Bayesian non-parametric models and inference for sparse and hierarchical latent structure (2012) pdf
PhD Thesis, University of Cambridge
Supervisor: Zoubin Ghahramani - Statistical tools for ulta-deep pyrosequencing of fast evolving viruses (2008) pdf
MSc Bioinformatics and Systems Biology, Imperial College London, Individual Project
Supervisor: Professor Susan Holmes, Stanford University - SBML-ABC: a package for data simulation, parameter inference and model selection, Group Report (2008) pdf
MSc Bioinformatics and Systems Biology, Imperial College London, Group Project
Supervisor: Professor Michael Stumpf
Presentations
- Properties of Bayesian nonparametric models and priors over trees. Guest lecture as part of Matt Hoffman's STAT300 class, summer 2013.
- Diffusion trees as priors. This was a talk I gave about the Dirichlet diffusion tree and Pitman Yor diffusion tree at Collegio Carlo Alberto.
- Variational methods for nonparametric Bayesian models
I gave a brief presentation at Microsoft Research summarising some attempts to use variational inference in nonparametric, particularly Dirichlet Process based, models. The slides are here.